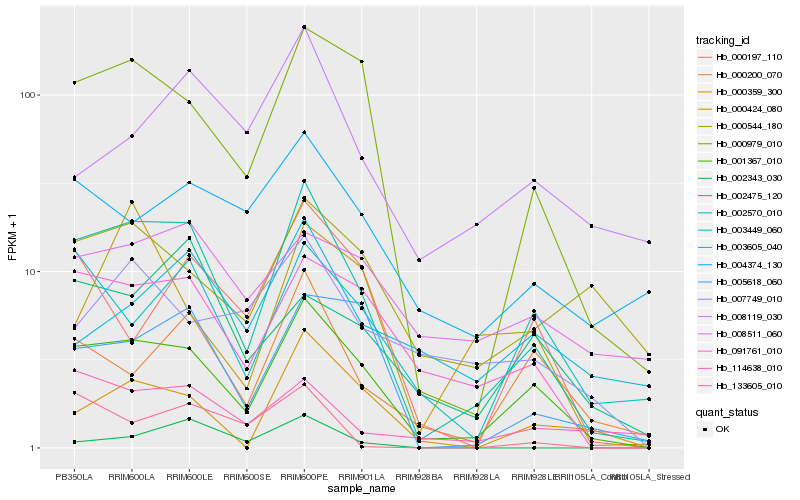
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002570_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 2 |
Hb_001367_010 |
0.1203287302 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 3 |
Hb_000979_010 |
0.1827190272 |
- |
- |
PREDICTED: uncharacterized protein LOC105128288 [Populus euphratica] |
| 4 |
Hb_091761_010 |
0.2029577291 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 5 |
Hb_005618_060 |
0.2233308839 |
transcription factor |
TF Family: bHLH |
hypothetical protein RCOM_1343120 [Ricinus communis] |
| 6 |
Hb_000200_070 |
0.2331588657 |
- |
- |
PREDICTED: CDP-diacylglycerol--serine O-phosphatidyltransferase 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_114638_010 |
0.2458957335 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_003449_060 |
0.2482893914 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 9 |
Hb_000197_110 |
0.2527491321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000359_300 |
0.2534498968 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Zea mays] |
| 11 |
Hb_002343_030 |
0.2535510225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004374_130 |
0.2555109289 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 13 |
Hb_002475_120 |
0.259482829 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 14 |
Hb_133605_010 |
0.2616133341 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 15 |
Hb_000544_180 |
0.2725598504 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X1 [Jatropha curcas] |
| 16 |
Hb_008511_060 |
0.2730163897 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 17 |
Hb_008119_030 |
0.2734563205 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 18 |
Hb_007749_010 |
0.2789229449 |
- |
- |
- |
| 19 |
Hb_000424_080 |
0.2816735363 |
- |
- |
PREDICTED: endoglucanase 16 [Jatropha curcas] |
| 20 |
Hb_003605_040 |
0.2822345738 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 isoform X1 [Jatropha curcas] |