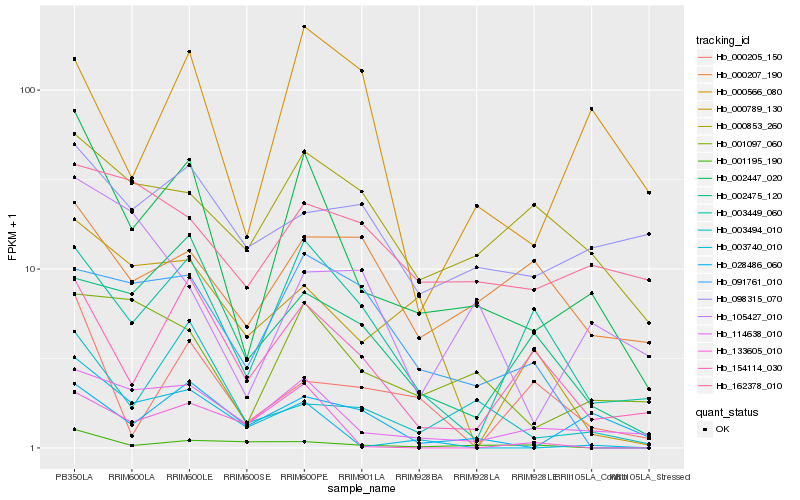
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002447_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_114638_010 |
0.201744148 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001097_060 |
0.2384464088 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8 [Jatropha curcas] |
| 4 |
Hb_154114_030 |
0.2399739009 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_003449_060 |
0.2481366777 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 6 |
Hb_003740_010 |
0.2559319876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003494_010 |
0.2605766229 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |
| 8 |
Hb_028486_060 |
0.2735839273 |
- |
- |
PREDICTED: putative germin-like protein 9-2 [Jatropha curcas] |
| 9 |
Hb_000853_260 |
0.2748608946 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 10 |
Hb_000789_130 |
0.2888402156 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 11 |
Hb_002475_120 |
0.2892952832 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 12 |
Hb_001195_190 |
0.2896964892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000566_080 |
0.2930921586 |
- |
- |
- |
| 14 |
Hb_091761_010 |
0.2964856773 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 15 |
Hb_162378_010 |
0.2965626741 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 16 |
Hb_000205_150 |
0.2981707216 |
- |
- |
PREDICTED: uncharacterized protein LOC105647752 [Jatropha curcas] |
| 17 |
Hb_098315_070 |
0.2994156216 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 18 |
Hb_105427_010 |
0.3031697715 |
- |
- |
- |
| 19 |
Hb_133605_010 |
0.3033885175 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_000207_190 |
0.3053641672 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |