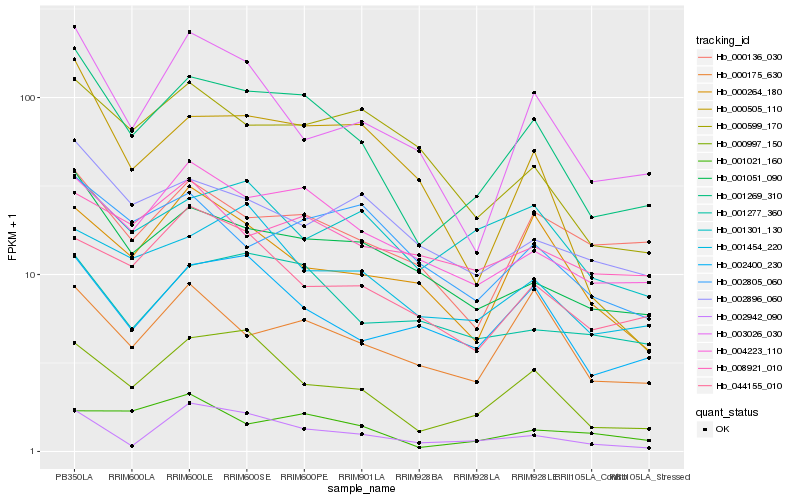
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_310 |
0.0 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 2 |
Hb_000997_150 |
0.1289199966 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_004223_110 |
0.1328424576 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 4 |
Hb_001051_090 |
0.1345976182 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 5 |
Hb_002400_230 |
0.1496984948 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 6 |
Hb_000175_630 |
0.1524135028 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g11310, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003026_030 |
0.1557490101 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 8 |
Hb_001454_220 |
0.1573030889 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 9 |
Hb_002896_060 |
0.1593204907 |
- |
- |
PREDICTED: synaptotagmin-2 [Jatropha curcas] |
| 10 |
Hb_000136_030 |
0.1623446424 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 11 |
Hb_001277_360 |
0.1626327022 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 12 |
Hb_044155_010 |
0.1643321025 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002805_060 |
0.1675070945 |
- |
- |
PREDICTED: signal peptide peptidase [Jatropha curcas] |
| 14 |
Hb_000505_110 |
0.1686420615 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 15 |
Hb_000264_180 |
0.1708013601 |
- |
- |
PREDICTED: uncharacterized protein LOC100777318 [Glycine max] |
| 16 |
Hb_002942_090 |
0.1712889911 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 17 |
Hb_001301_130 |
0.1720525161 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 18 |
Hb_001021_160 |
0.1738469859 |
- |
- |
PREDICTED: homeobox protein LUMINIDEPENDENS [Jatropha curcas] |
| 19 |
Hb_000599_170 |
0.1745975567 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_008921_010 |
0.1747097798 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |