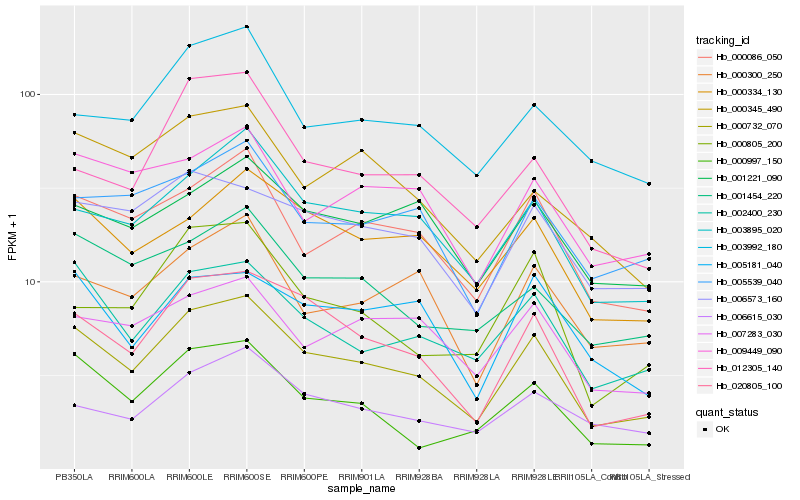
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_070 |
0.0 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 2 |
Hb_020805_100 |
0.0816371544 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 3 |
Hb_000086_050 |
0.092705343 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 4 |
Hb_003895_020 |
0.0933015306 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002400_230 |
0.096554889 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 6 |
Hb_000334_130 |
0.1062609281 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 7 |
Hb_006573_160 |
0.1131122123 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |
| 8 |
Hb_005539_040 |
0.1141109835 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000300_250 |
0.114398204 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001454_220 |
0.1145871449 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 11 |
Hb_000997_150 |
0.1176030655 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_006615_030 |
0.1178048863 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 13 |
Hb_009449_090 |
0.1178061084 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007283_030 |
0.1186667489 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 15 |
Hb_005181_040 |
0.118961215 |
- |
- |
PREDICTED: MAP kinase kinase MKK1/SSP32-like [Jatropha curcas] |
| 16 |
Hb_003992_180 |
0.1192244756 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000345_490 |
0.1192629225 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 18 |
Hb_001221_090 |
0.1204510513 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000805_200 |
0.1209439746 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_012305_140 |
0.1214303332 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |