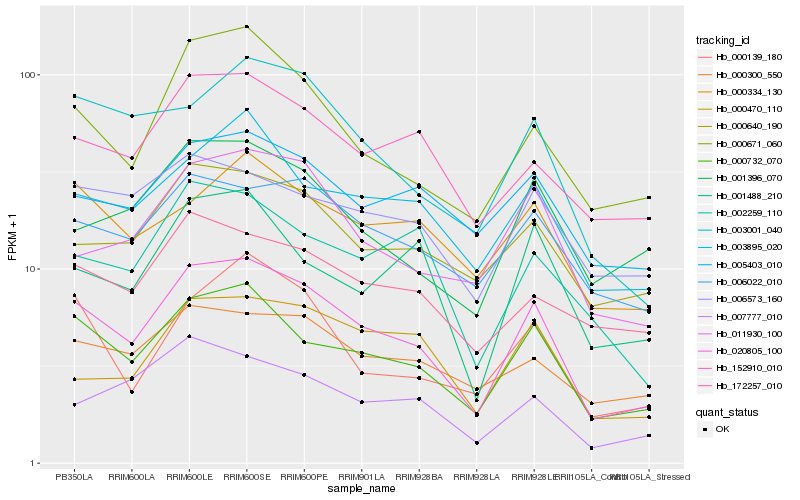
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020805_100 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 2 |
Hb_000732_070 |
0.0816371544 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 3 |
Hb_000671_060 |
0.1169274684 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_003895_020 |
0.1216062564 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003001_040 |
0.1230418176 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 6 |
Hb_006022_010 |
0.1234855086 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 7 |
Hb_152910_010 |
0.1257732717 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |
| 8 |
Hb_000640_190 |
0.1271421299 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011930_100 |
0.127166127 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 10 |
Hb_000139_180 |
0.1317646713 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 11 |
Hb_007777_010 |
0.1319096012 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Tarenaya hassleriana] |
| 12 |
Hb_001396_070 |
0.1331565302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000300_550 |
0.1338020074 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 14 |
Hb_002259_110 |
0.1342906472 |
- |
- |
PREDICTED: uncharacterized protein LOC105630166 [Jatropha curcas] |
| 15 |
Hb_000334_130 |
0.1348916922 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 16 |
Hb_000470_110 |
0.1355142979 |
- |
- |
PREDICTED: probable LIM domain-containing serine/threonine-protein kinase DDB_G0287001 [Jatropha curcas] |
| 17 |
Hb_005403_010 |
0.1355899884 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 18 |
Hb_001488_210 |
0.1375767555 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 19 |
Hb_172257_010 |
0.1379745249 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 20 |
Hb_006573_160 |
0.1381388115 |
- |
- |
PREDICTED: dnaJ protein ERDJ2A-like [Jatropha curcas] |