| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
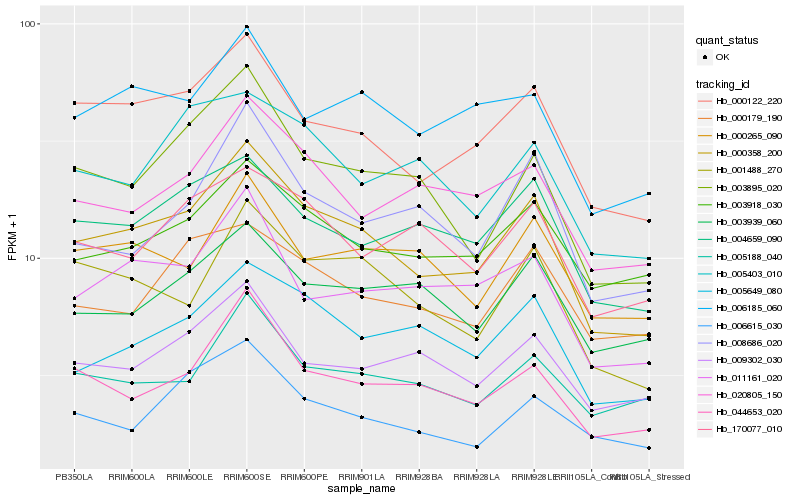
Hb_000358_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 2 |
Hb_000122_220 |
0.0699556107 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 3 |
Hb_001488_270 |
0.0900202344 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 4 |
Hb_003895_020 |
0.0921148014 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 5 |
Hb_044653_020 |
0.0921511993 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 6 |
Hb_005649_080 |
0.09445042 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 7 |
Hb_004659_090 |
0.0948982417 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 8 |
Hb_003918_030 |
0.0963376968 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_020805_150 |
0.0975782216 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_011161_020 |
0.0983283381 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 11 |
Hb_006185_060 |
0.0991211707 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 12 |
Hb_009302_030 |
0.1027282918 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 13 |
Hb_000265_090 |
0.10534534 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 14 |
Hb_005188_040 |
0.1058667708 |
- |
- |
PREDICTED: protein furry homolog-like [Nicotiana sylvestris] |
| 15 |
Hb_006615_030 |
0.1059701467 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 16 |
Hb_003939_060 |
0.1059781203 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_008686_020 |
0.1060524212 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000179_190 |
0.106608807 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 19 |
Hb_170077_010 |
0.1067322954 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005403_010 |
0.1073125065 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |