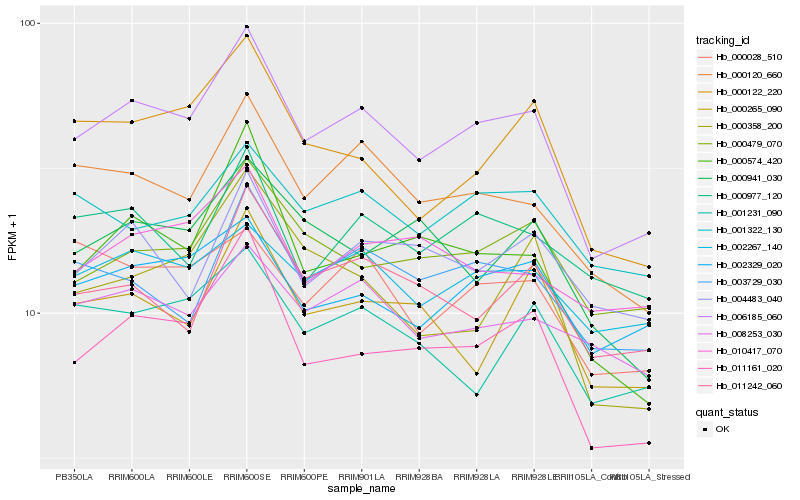
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006185_060 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 2 |
Hb_011161_020 |
0.0638101604 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 3 |
Hb_000120_660 |
0.0760644874 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 4 |
Hb_000122_220 |
0.0869651841 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 5 |
Hb_000574_420 |
0.0873841271 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 6 |
Hb_002329_020 |
0.088359121 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 7 |
Hb_000479_070 |
0.0892398156 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 8 |
Hb_003729_030 |
0.090549887 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 9 |
Hb_002267_140 |
0.0944399857 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 10 |
Hb_011242_060 |
0.0960322438 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 11 |
Hb_000028_510 |
0.0964270603 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 12 |
Hb_008253_030 |
0.0979125168 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 13 |
Hb_000977_120 |
0.0980459161 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001322_130 |
0.0980879822 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 15 |
Hb_000358_200 |
0.0991211707 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 16 |
Hb_010417_070 |
0.0995800461 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001231_090 |
0.0995972393 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004483_040 |
0.0997765153 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 19 |
Hb_000265_090 |
0.0999664441 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 20 |
Hb_000941_030 |
0.1006847811 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |