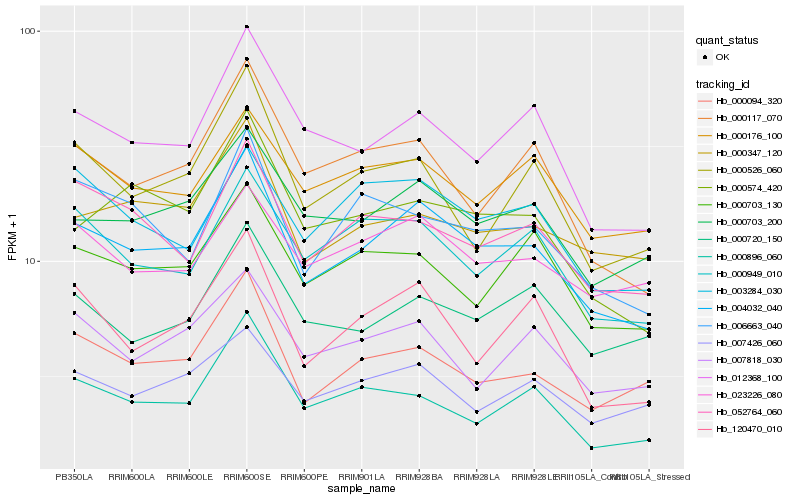
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_040 |
0.0 |
- |
- |
unknown [Medicago truncatula] |
| 2 |
Hb_000703_200 |
0.085379559 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_012368_100 |
0.0877055513 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 4 |
Hb_000094_320 |
0.0893554724 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 5 |
Hb_120470_010 |
0.0933939128 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_000117_070 |
0.0943120359 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 7 |
Hb_000574_420 |
0.0951677488 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 8 |
Hb_052764_060 |
0.095795932 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 9 |
Hb_000949_010 |
0.0962696441 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 10 |
Hb_000347_120 |
0.0968789655 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 11 |
Hb_023226_080 |
0.0983061274 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000896_060 |
0.0988032168 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_006663_040 |
0.0989959908 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003284_030 |
0.1000490348 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000703_130 |
0.1002698099 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 16 |
Hb_000176_100 |
0.1007447849 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 17 |
Hb_007426_060 |
0.1009298876 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_007818_030 |
0.1015625449 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 19 |
Hb_000526_060 |
0.1016873517 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 20 |
Hb_000720_150 |
0.1030526942 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X3 [Jatropha curcas] |