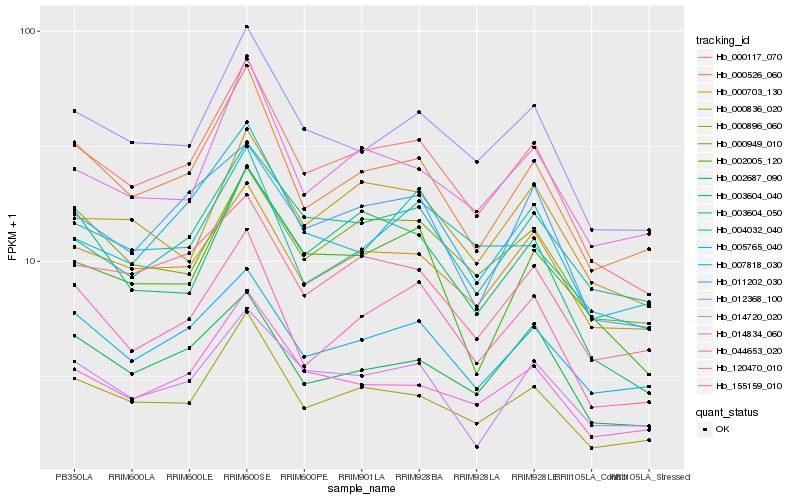
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000117_070 |
0.0 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 2 |
Hb_000896_060 |
0.0597894729 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_012368_100 |
0.0623235384 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 4 |
Hb_000526_060 |
0.0650852722 |
- |
- |
PREDICTED: clustered mitochondria protein [Jatropha curcas] |
| 5 |
Hb_044653_020 |
0.0737308296 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 6 |
Hb_120470_010 |
0.07514026 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 7 |
Hb_000703_130 |
0.0775472114 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 8 |
Hb_155159_010 |
0.0797607162 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 9 |
Hb_011202_030 |
0.0821371163 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 10 |
Hb_003604_050 |
0.0846255157 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000949_010 |
0.0848877206 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 12 |
Hb_014834_060 |
0.0851244533 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007818_030 |
0.0860119644 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like [Jatropha curcas] |
| 14 |
Hb_002687_090 |
0.0869002536 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_014720_020 |
0.088220501 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 16 |
Hb_002005_120 |
0.0884440447 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 17 |
Hb_005765_040 |
0.0899606045 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 18 |
Hb_000836_020 |
0.0904025921 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 19 |
Hb_003604_040 |
0.0917849072 |
- |
- |
Serine/threonine-protein kinase sepA [Morus notabilis] |
| 20 |
Hb_004032_040 |
0.0943120359 |
- |
- |
unknown [Medicago truncatula] |