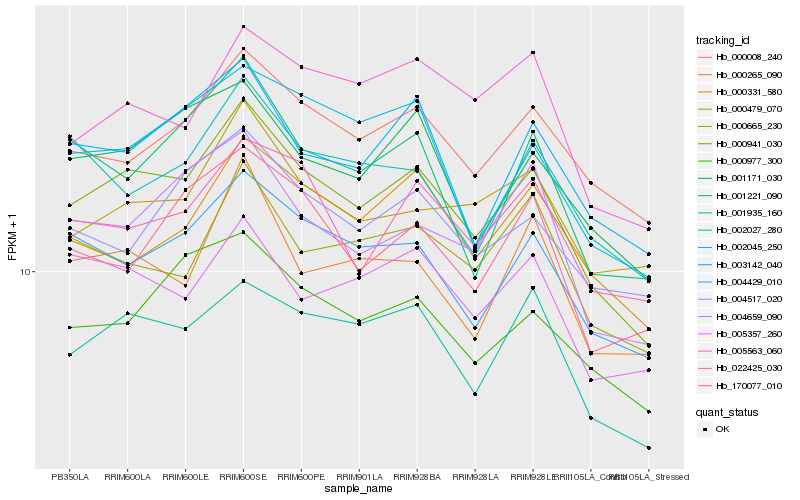
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000941_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646531 [Jatropha curcas] |
| 2 |
Hb_001935_160 |
0.0643922007 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 3 |
Hb_004429_010 |
0.0687676548 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_001171_030 |
0.074945294 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 5 |
Hb_000008_240 |
0.0760452619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000977_300 |
0.0775944296 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 7 |
Hb_004659_090 |
0.0785325041 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 8 |
Hb_003142_040 |
0.0795770149 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 9 |
Hb_022425_030 |
0.0812047521 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 10 |
Hb_002045_250 |
0.0826651566 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000265_090 |
0.0836971475 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 12 |
Hb_001221_090 |
0.0848097482 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000665_230 |
0.0848716627 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 14 |
Hb_005563_060 |
0.0858831785 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 15 |
Hb_005357_260 |
0.0861728273 |
- |
- |
PREDICTED: polyadenylation and cleavage factor homolog 4 [Jatropha curcas] |
| 16 |
Hb_000331_580 |
0.0862098803 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 17 |
Hb_004517_020 |
0.0865237096 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 18 |
Hb_002027_280 |
0.086830679 |
- |
- |
PREDICTED: sister-chromatid cohesion protein 3 [Jatropha curcas] |
| 19 |
Hb_170077_010 |
0.0876324105 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000479_070 |
0.0876988964 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |