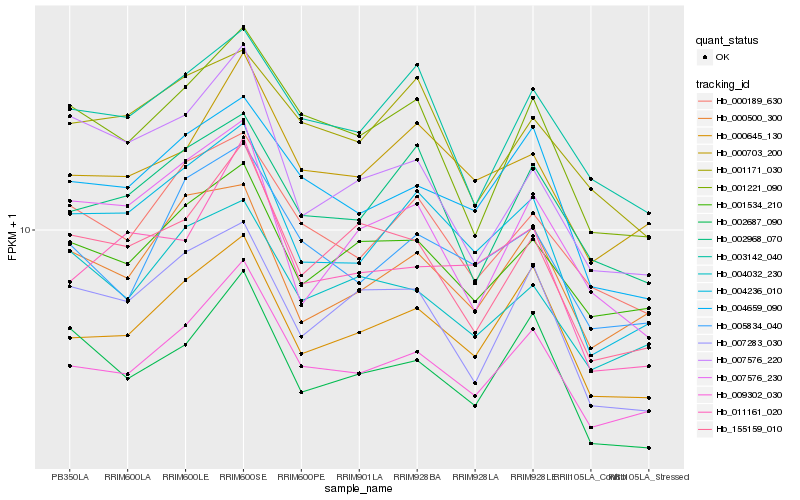
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004236_010 |
0.0 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 2 |
Hb_000645_130 |
0.0772637731 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 3 |
Hb_009302_030 |
0.0776712132 |
- |
- |
hypothetical protein RCOM_1176340 [Ricinus communis] |
| 4 |
Hb_000500_300 |
0.0778067937 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 5 |
Hb_004659_090 |
0.0797881424 |
- |
- |
PREDICTED: uncharacterized protein LOC105629935 [Jatropha curcas] |
| 6 |
Hb_000189_630 |
0.0800917419 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_007283_030 |
0.0866367386 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |
| 8 |
Hb_000703_200 |
0.0896876222 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_007576_230 |
0.0897408662 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 10 |
Hb_002968_070 |
0.0904977853 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 11 |
Hb_001534_210 |
0.090902509 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011161_020 |
0.0910569798 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 13 |
Hb_005834_040 |
0.0920786079 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 14 |
Hb_002687_090 |
0.0923592217 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_003142_040 |
0.0943366763 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 16 |
Hb_007576_220 |
0.0953982769 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 17 |
Hb_001221_090 |
0.0961355279 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001171_030 |
0.0972296068 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 19 |
Hb_155159_010 |
0.0978054575 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 1 [Jatropha curcas] |
| 20 |
Hb_004032_230 |
0.0978125283 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |