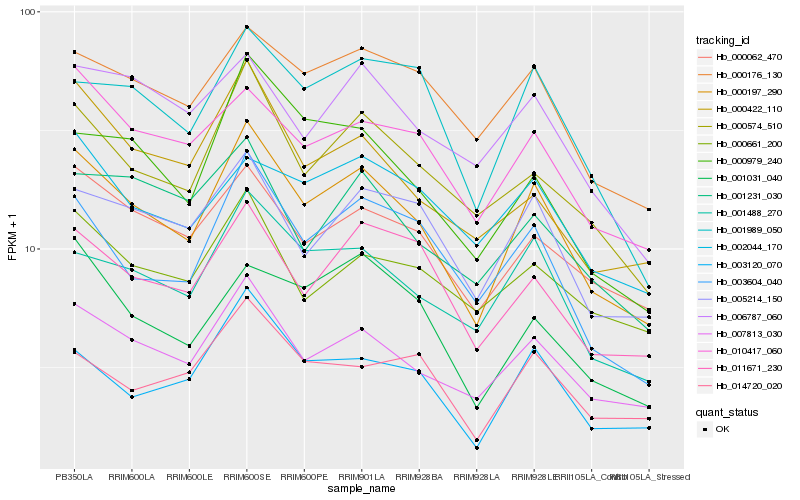
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_290 |
0.0 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 2 |
Hb_007813_030 |
0.070361864 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 3 |
Hb_001488_270 |
0.0867542777 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 4 |
Hb_000574_510 |
0.0887753403 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000062_470 |
0.0933520717 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 6 |
Hb_003604_040 |
0.0952706826 |
- |
- |
Serine/threonine-protein kinase sepA [Morus notabilis] |
| 7 |
Hb_003120_070 |
0.0957574497 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_005214_150 |
0.0963989904 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 9 |
Hb_010417_060 |
0.0982520168 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 10 |
Hb_011671_230 |
0.0985058375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000176_130 |
0.1006293511 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Jatropha curcas] |
| 12 |
Hb_001989_050 |
0.1036765993 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 13 |
Hb_001031_040 |
0.106010827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000979_240 |
0.107984195 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 15 |
Hb_000422_110 |
0.1085403222 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 16 |
Hb_006787_060 |
0.1108090801 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000661_200 |
0.1110032809 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 18 |
Hb_014720_020 |
0.1115094044 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 19 |
Hb_001231_030 |
0.1124514511 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 20 |
Hb_002044_170 |
0.1136718187 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |