| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
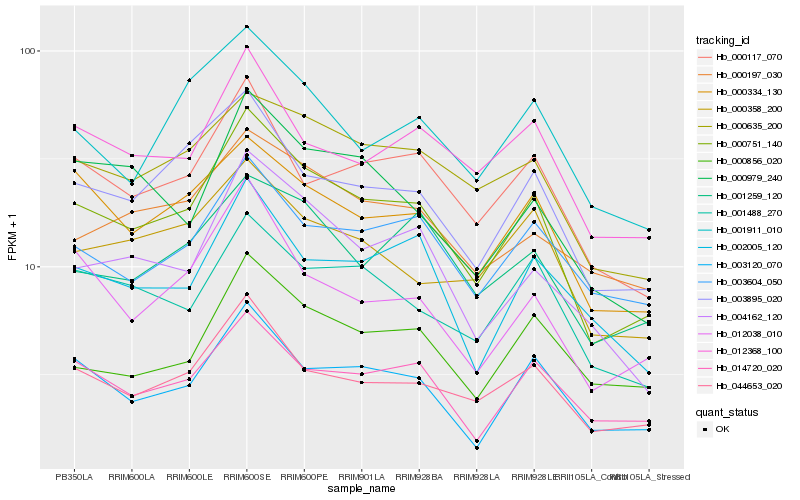
Hb_000751_140 |
0.0 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 2 |
Hb_044653_020 |
0.0880127136 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 3 |
Hb_003895_020 |
0.0903166048 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003120_070 |
0.0919069142 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000334_130 |
0.0967681446 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 6 |
Hb_000117_070 |
0.0968108158 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 7 |
Hb_000635_200 |
0.0974460214 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 8 |
Hb_004162_120 |
0.0983980592 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 41 homolog [Populus euphratica] |
| 9 |
Hb_014720_020 |
0.1030989017 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 10 |
Hb_001488_270 |
0.1041108642 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 11 |
Hb_012038_010 |
0.1050736218 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 12 |
Hb_012368_100 |
0.1064950349 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 13 |
Hb_003604_050 |
0.1079222998 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_001911_010 |
0.1087500149 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 15 |
Hb_000358_200 |
0.1094709254 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 16 |
Hb_001259_120 |
0.1100719212 |
- |
- |
hypothetical protein EUGRSUZ_C043222, partial [Eucalyptus grandis] |
| 17 |
Hb_000979_240 |
0.1104007689 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 18 |
Hb_000856_020 |
0.1108253403 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 19 |
Hb_000197_030 |
0.1108639316 |
- |
- |
hypothetical protein RCOM_1447500 [Ricinus communis] |
| 20 |
Hb_002005_120 |
0.1109439338 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |