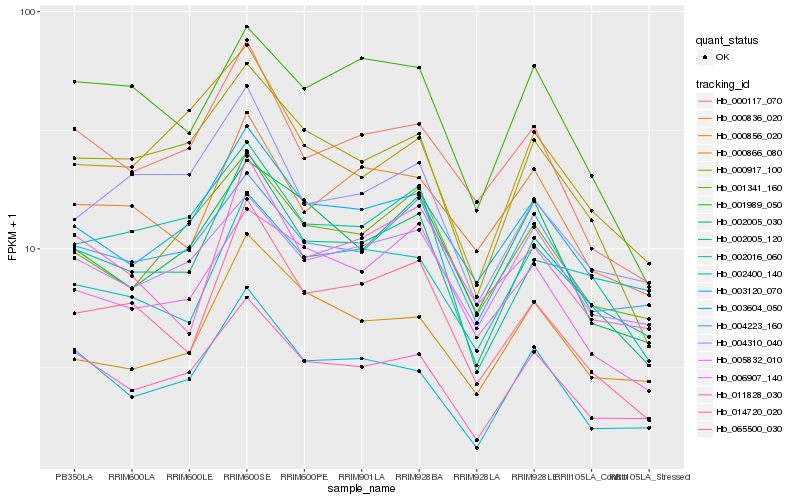
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_120 |
0.0 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 2 |
Hb_014720_020 |
0.0712033526 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 3 |
Hb_065500_030 |
0.0717536056 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 4 |
Hb_000917_100 |
0.0789460124 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 5 |
Hb_003604_050 |
0.0825434487 |
- |
- |
PREDICTED: MAP3K epsilon protein kinase 1-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_002016_060 |
0.087611129 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 7 |
Hb_003120_070 |
0.087784669 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000117_070 |
0.0884440447 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 9 |
Hb_001341_160 |
0.090385397 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002400_140 |
0.0950213727 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_006907_140 |
0.09605596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004223_160 |
0.0972304476 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004310_040 |
0.0994931648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002005_030 |
0.1007964288 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 15 |
Hb_000836_020 |
0.1030888342 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 16 |
Hb_005832_010 |
0.1035265498 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001989_050 |
0.1041229726 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 5 [Jatropha curcas] |
| 18 |
Hb_011828_030 |
0.1049340752 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 19 |
Hb_000866_080 |
0.1051207227 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000856_020 |
0.1054071391 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |