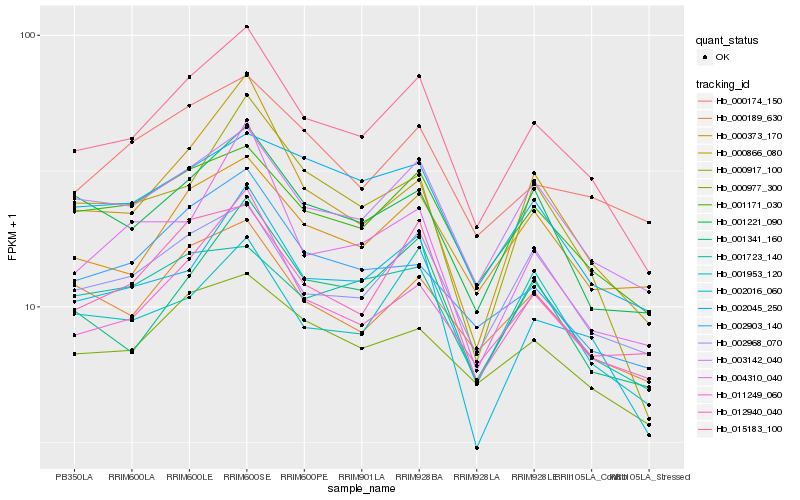
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015183_100 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360 [Jatropha curcas] |
| 2 |
Hb_003142_040 |
0.0696706815 |
- |
- |
Sf3a3 [Gossypium arboreum] |
| 3 |
Hb_001171_030 |
0.0701804542 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like [Jatropha curcas] |
| 4 |
Hb_011249_060 |
0.0733792208 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 5 |
Hb_002968_070 |
0.0736450742 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 4b isoform X2 [Jatropha curcas] |
| 6 |
Hb_012940_040 |
0.0756090485 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 7 |
Hb_001341_160 |
0.0779468228 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000866_080 |
0.0785017261 |
- |
- |
PREDICTED: signal peptide peptidase-like 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000977_300 |
0.0814084459 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable E3 ubiquitin-protein ligase ARI8 [Malus domestica] |
| 10 |
Hb_001221_090 |
0.0814162825 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002016_060 |
0.081887784 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 12 |
Hb_000189_630 |
0.0837396457 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 13 |
Hb_002903_140 |
0.0847102128 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 14 |
Hb_002045_250 |
0.0878331905 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000373_170 |
0.0885295461 |
- |
- |
PREDICTED: exocyst complex component SEC5A-like [Jatropha curcas] |
| 16 |
Hb_000174_150 |
0.089086938 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 17 |
Hb_004310_040 |
0.0898460482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001953_120 |
0.0904155121 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000917_100 |
0.0920659003 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 20 |
Hb_001723_140 |
0.0945394109 |
- |
- |
PREDICTED: importin beta-like SAD2 homolog isoform X2 [Jatropha curcas] |