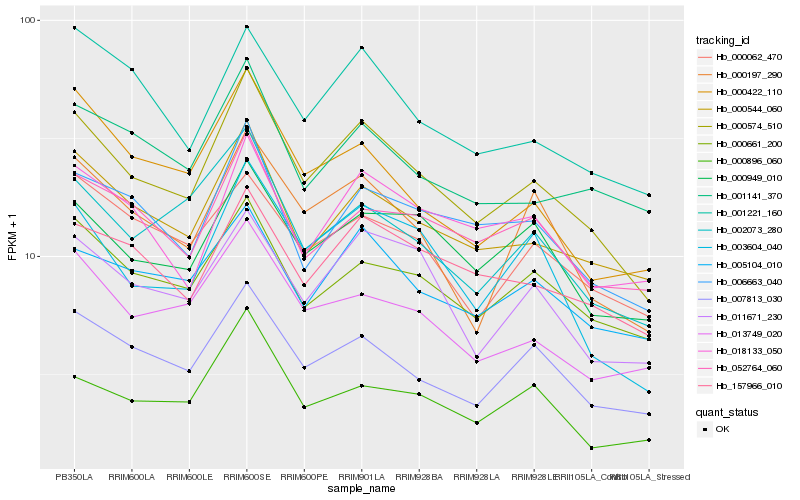
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_510 |
0.0 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000544_060 |
0.0790504889 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 3 |
Hb_007813_030 |
0.0797653505 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 4 |
Hb_003604_040 |
0.084892427 |
- |
- |
Serine/threonine-protein kinase sepA [Morus notabilis] |
| 5 |
Hb_000661_200 |
0.0873464048 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000197_290 |
0.0887753403 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 7 |
Hb_011671_230 |
0.0895993316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006663_040 |
0.0899155966 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_013749_020 |
0.0905421033 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_000949_010 |
0.0912838202 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF2 [Jatropha curcas] |
| 11 |
Hb_005104_010 |
0.0916749261 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 12 |
Hb_157966_010 |
0.091837293 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 13 |
Hb_002073_280 |
0.092558539 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001221_160 |
0.0946524179 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 15 |
Hb_052764_060 |
0.0960384673 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000422_110 |
0.0972839405 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 17 |
Hb_000896_060 |
0.0994652639 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001141_370 |
0.0995163034 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 19 |
Hb_018133_050 |
0.1004595617 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 1 [Jatropha curcas] |
| 20 |
Hb_000062_470 |
0.1008083622 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |