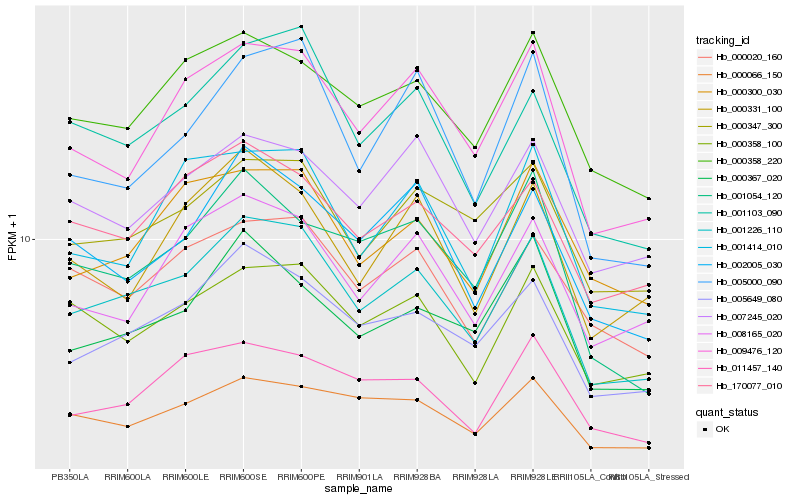
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001226_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638106 [Jatropha curcas] |
| 2 |
Hb_005649_080 |
0.0638216325 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 3 |
Hb_001103_090 |
0.0753642368 |
- |
- |
PREDICTED: patellin-3 [Populus euphratica] |
| 4 |
Hb_009476_120 |
0.0778395592 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 5 |
Hb_000358_220 |
0.0816876715 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 6 |
Hb_170077_010 |
0.0823101655 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000358_100 |
0.0838486236 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 8 |
Hb_000020_160 |
0.0848264623 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 9 |
Hb_000347_300 |
0.0860172033 |
- |
- |
PREDICTED: cation/calcium exchanger 4-like [Jatropha curcas] |
| 10 |
Hb_001414_010 |
0.0885120709 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 11 |
Hb_008165_020 |
0.0892032286 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000300_030 |
0.0893485423 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 13 |
Hb_002005_030 |
0.0901785539 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 14 |
Hb_011457_140 |
0.091633161 |
- |
- |
PREDICTED: uncharacterized protein LOC105649947 [Jatropha curcas] |
| 15 |
Hb_000367_020 |
0.0924789848 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 16 |
Hb_007245_020 |
0.0951783024 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 17 |
Hb_005000_090 |
0.0952623478 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 18 |
Hb_001054_120 |
0.0963143156 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 19 |
Hb_000066_150 |
0.0968760153 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 20 |
Hb_000331_100 |
0.097001456 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |