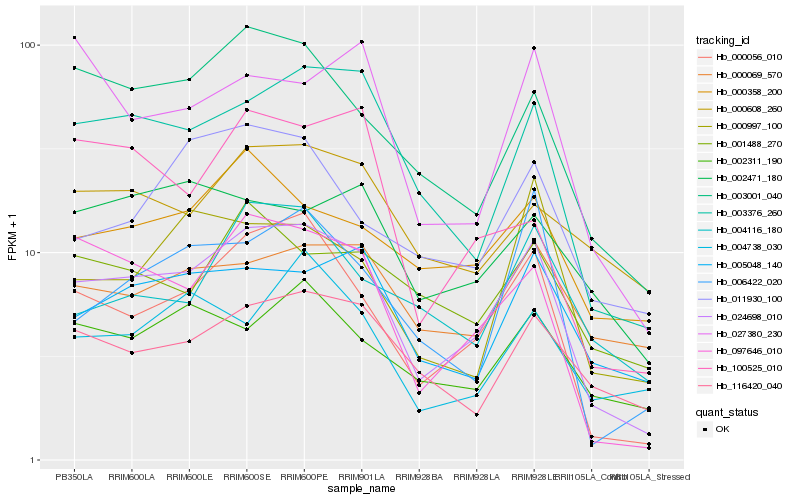
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024698_010 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46 [Jatropha curcas] |
| 2 |
Hb_000056_010 |
0.100051406 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 3 |
Hb_003376_260 |
0.117502098 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 4 |
Hb_097646_010 |
0.1178502156 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 5 |
Hb_003001_040 |
0.1346822824 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 6 |
Hb_005048_140 |
0.1496169816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_116420_040 |
0.1499715456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002311_190 |
0.1617377504 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 9 |
Hb_000358_200 |
0.1617954132 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 10 |
Hb_006422_020 |
0.1648917457 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 11 |
Hb_000069_570 |
0.1658464425 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000997_100 |
0.1682690666 |
- |
- |
PREDICTED: uncharacterized protein LOC105632094 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000608_260 |
0.1685630575 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 14 |
Hb_001488_270 |
0.1695712994 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 15 |
Hb_004116_180 |
0.1707256941 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 16 |
Hb_002471_180 |
0.1707994021 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 17 |
Hb_011930_100 |
0.1710608438 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 18 |
Hb_004738_030 |
0.172000625 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 19 |
Hb_027380_230 |
0.1721410002 |
- |
- |
PREDICTED: CSC1-like protein ERD4 [Jatropha curcas] |
| 20 |
Hb_100525_010 |
0.177080898 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |