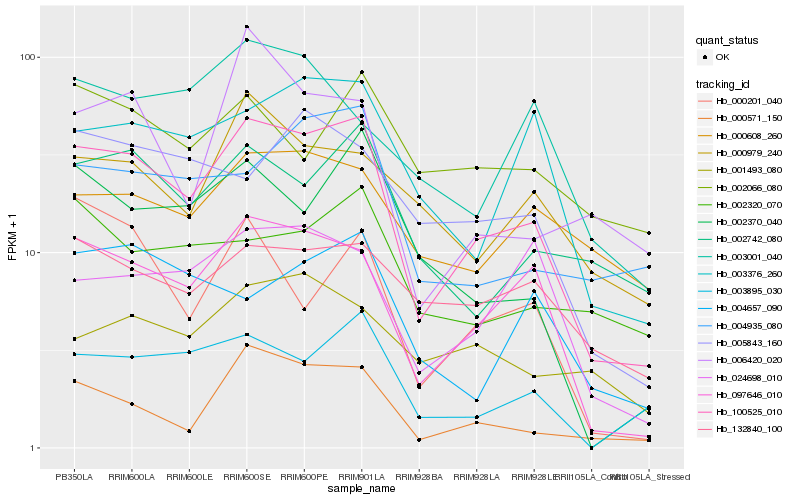
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100525_010 |
0.0 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 2 |
Hb_097646_010 |
0.1176646192 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 3 |
Hb_000571_150 |
0.1570308767 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_003895_030 |
0.1623204093 |
- |
- |
- |
| 5 |
Hb_002742_080 |
0.1688694315 |
- |
- |
unknown [Populus trichocarpa] |
| 6 |
Hb_002370_040 |
0.171811554 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 7 |
Hb_003376_260 |
0.1718405309 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 8 |
Hb_024698_010 |
0.177080898 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46 [Jatropha curcas] |
| 9 |
Hb_004935_080 |
0.1777917728 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 10 |
Hb_132840_100 |
0.1808068285 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 11 |
Hb_000979_240 |
0.1818031608 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 12 |
Hb_005843_160 |
0.1847534021 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 13 |
Hb_000608_260 |
0.1885825274 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 14 |
Hb_002320_070 |
0.1905082956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004657_090 |
0.1930433254 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 16 |
Hb_006420_020 |
0.1935893718 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 17 |
Hb_000201_040 |
0.1936448508 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 18 |
Hb_001493_080 |
0.194740114 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 19 |
Hb_003001_040 |
0.1949403971 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 20 |
Hb_002066_080 |
0.1991068789 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |