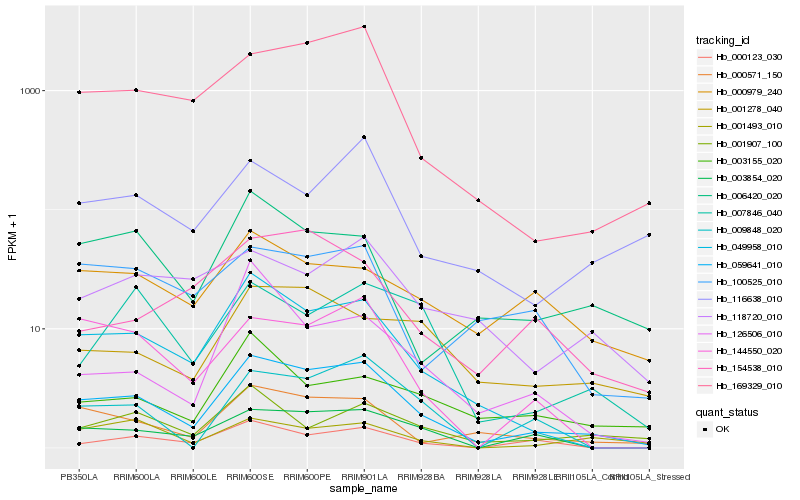
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_059641_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 2 |
Hb_049958_010 |
0.1531529181 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 3 |
Hb_000571_150 |
0.1691121305 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_169329_010 |
0.1725901389 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003854_020 |
0.1758233566 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_144550_020 |
0.1979709485 |
- |
- |
- |
| 7 |
Hb_000123_030 |
0.2021822978 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 8 |
Hb_001278_040 |
0.2028058598 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_006420_020 |
0.2059013963 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 10 |
Hb_009848_020 |
0.2065838028 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 11 |
Hb_116638_010 |
0.2129244343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001907_100 |
0.2174215695 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 13 |
Hb_003155_020 |
0.2179607657 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 14 |
Hb_100525_010 |
0.2187777428 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 15 |
Hb_000979_240 |
0.2197746612 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 16 |
Hb_001493_010 |
0.22015924 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 17 |
Hb_154538_010 |
0.2209443272 |
- |
- |
- |
| 18 |
Hb_007846_040 |
0.222039601 |
- |
- |
PREDICTED: uncharacterized protein LOC105646691 [Jatropha curcas] |
| 19 |
Hb_118720_010 |
0.2224200427 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 20 |
Hb_126506_010 |
0.224093863 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |