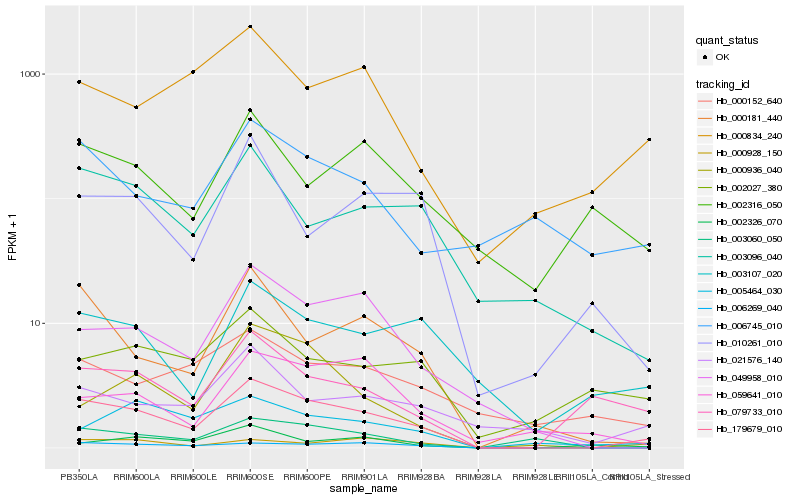
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179679_010 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_006269_040 |
0.192675977 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 3 |
Hb_049958_010 |
0.2000846969 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_000181_440 |
0.2039290982 |
- |
- |
- |
| 5 |
Hb_079733_010 |
0.2111101087 |
- |
- |
- |
| 6 |
Hb_003096_040 |
0.2167212269 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 7 |
Hb_003107_020 |
0.2204638339 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 8 |
Hb_021576_140 |
0.2245097302 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 9 |
Hb_003060_050 |
0.2259286734 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 10 |
Hb_010261_010 |
0.2310508828 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000928_150 |
0.2311207028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000834_240 |
0.2334220542 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 13 |
Hb_005464_030 |
0.2343857168 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_059641_010 |
0.2406963942 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_000152_640 |
0.2410490996 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 16 |
Hb_000936_040 |
0.2411555873 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 17 |
Hb_002027_380 |
0.2426462847 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 18 |
Hb_002316_050 |
0.2434528289 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002326_070 |
0.2461339074 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_006745_010 |
0.247351474 |
- |
- |
PREDICTED: uncharacterized protein LOC105640095 [Jatropha curcas] |