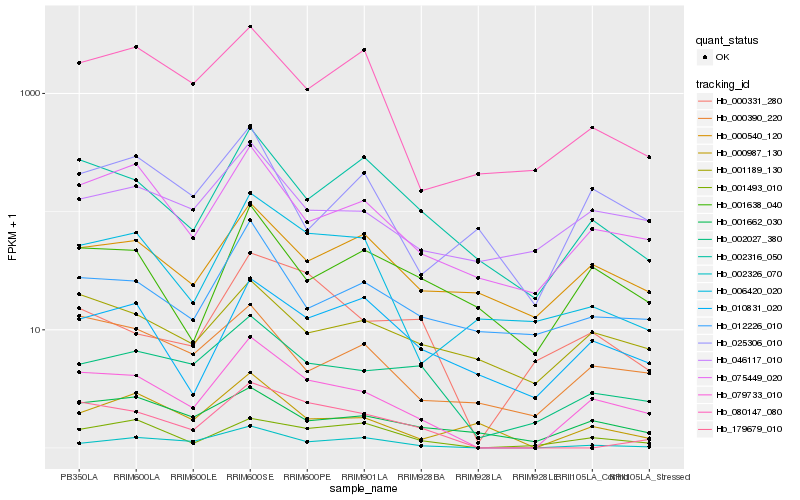
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079733_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_075449_020 |
0.1771956332 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 3 |
Hb_002027_380 |
0.1844615342 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 4 |
Hb_002326_070 |
0.1854632965 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001493_010 |
0.1924877596 |
- |
- |
triacylglycerol acidic lipase TAL4 [Ricinus communis] |
| 6 |
Hb_002316_050 |
0.1958143824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000390_220 |
0.1961011145 |
- |
- |
PREDICTED: uncharacterized protein LOC100244219 [Vitis vinifera] |
| 8 |
Hb_001662_030 |
0.2035310669 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 9 |
Hb_046117_010 |
0.2076692405 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 10 |
Hb_006420_020 |
0.2087923389 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_179679_010 |
0.2111101087 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 12 |
Hb_001638_040 |
0.2154824287 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 13 |
Hb_080147_080 |
0.216607781 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 14 |
Hb_000987_130 |
0.2183852928 |
- |
- |
- |
| 15 |
Hb_010831_020 |
0.2185922856 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 16 |
Hb_001189_130 |
0.2191937325 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 17 |
Hb_000540_120 |
0.2194641836 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 18 |
Hb_000331_280 |
0.2200743845 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 19 |
Hb_012226_010 |
0.2206909019 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 20 |
Hb_025306_010 |
0.2209717217 |
transcription factor |
TF Family: MYB-related |
syringolide-induced protein 1-3-1B [Populus trichocarpa] |