| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
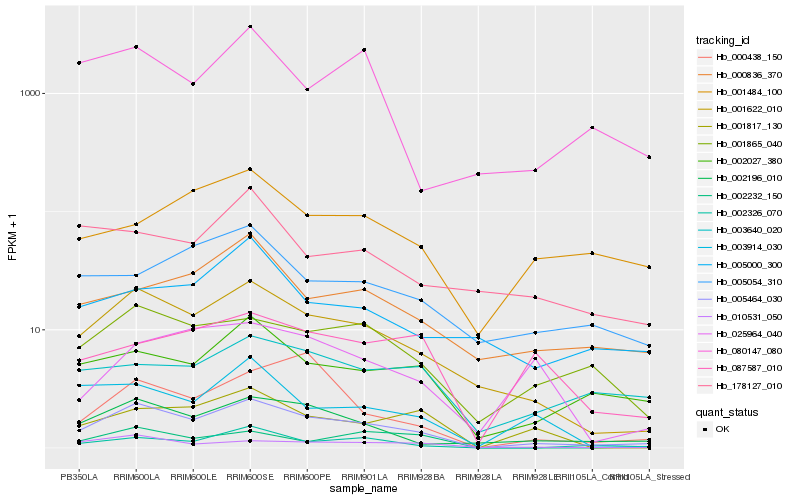
Hb_005464_030 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001622_010 |
0.1250993872 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_002196_010 |
0.1613157764 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: proline-rich receptor-like protein kinase PERK1 [Jatropha curcas] |
| 4 |
Hb_001865_040 |
0.1829290814 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_002027_380 |
0.1830258798 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase isoform X2 [Jatropha curcas] |
| 6 |
Hb_010531_050 |
0.1873513073 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002326_070 |
0.1925150121 |
- |
- |
ent-kaurene oxidase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000836_370 |
0.2080408292 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001817_130 |
0.2104686094 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 10 |
Hb_000438_150 |
0.2148300063 |
transcription factor |
TF Family: HMG |
PREDICTED: high mobility group B protein 7 [Jatropha curcas] |
| 11 |
Hb_003914_030 |
0.2160936928 |
- |
- |
- |
| 12 |
Hb_002232_150 |
0.2195360323 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 13 |
Hb_003640_020 |
0.2200360533 |
- |
- |
pelota, putative [Ricinus communis] |
| 14 |
Hb_005054_310 |
0.2236467163 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 15 |
Hb_080147_080 |
0.2243610596 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 16 |
Hb_025964_040 |
0.2248368052 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 17 |
Hb_005000_300 |
0.2256195724 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 18 |
Hb_087587_010 |
0.2268696935 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 19 |
Hb_001484_100 |
0.2304845293 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 20 |
Hb_178127_010 |
0.2325647585 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |