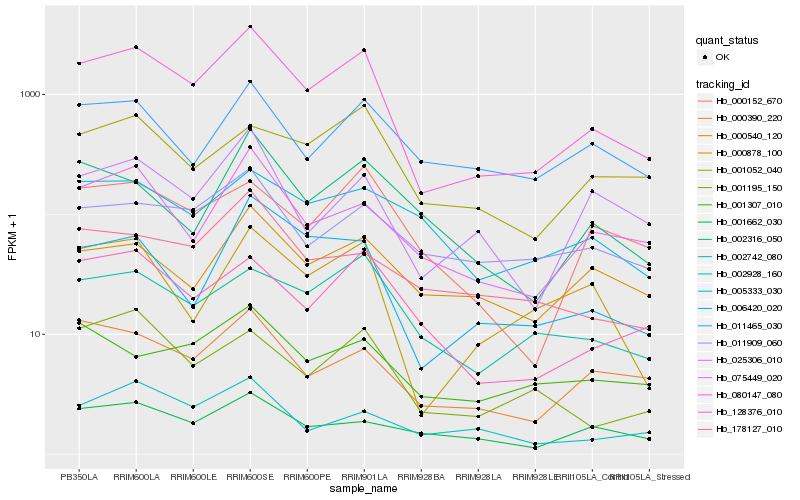
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080147_080 |
0.0 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 2 |
Hb_006420_020 |
0.1485442128 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 3 |
Hb_075449_020 |
0.1490750504 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 4 |
Hb_128376_010 |
0.1498047479 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000390_220 |
0.1503564744 |
- |
- |
PREDICTED: uncharacterized protein LOC100244219 [Vitis vinifera] |
| 6 |
Hb_000878_100 |
0.1549721032 |
- |
- |
PREDICTED: ABC transporter G family member 22 [Jatropha curcas] |
| 7 |
Hb_002742_080 |
0.1550864558 |
- |
- |
unknown [Populus trichocarpa] |
| 8 |
Hb_025306_010 |
0.1605807873 |
transcription factor |
TF Family: MYB-related |
syringolide-induced protein 1-3-1B [Populus trichocarpa] |
| 9 |
Hb_001195_150 |
0.1651669217 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 10 |
Hb_001662_030 |
0.1686301706 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 11 |
Hb_000152_670 |
0.1691272968 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 12 |
Hb_011909_060 |
0.1692866625 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 13 |
Hb_178127_010 |
0.1693783587 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 14 |
Hb_002928_160 |
0.1694914069 |
- |
- |
PREDICTED: U-box domain-containing protein 15 [Jatropha curcas] |
| 15 |
Hb_002316_050 |
0.1712778616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001052_040 |
0.1721938674 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_011465_030 |
0.1732153626 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 18 |
Hb_000540_120 |
0.1762664399 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 19 |
Hb_005333_030 |
0.1766212171 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 20 |
Hb_001307_010 |
0.176879269 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |