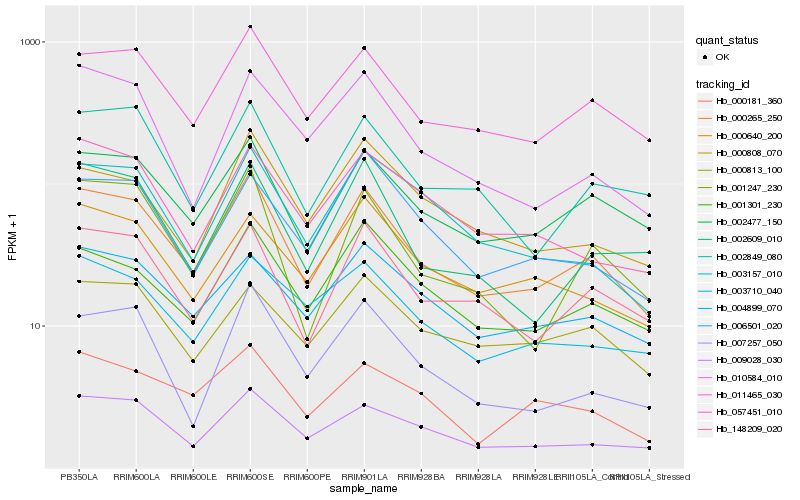
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000265_250 |
0.0 |
- |
- |
auxilin, putative [Ricinus communis] |
| 2 |
Hb_011465_030 |
0.0743271987 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_002477_150 |
0.083031228 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_148209_020 |
0.0864963889 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 5 |
Hb_009028_030 |
0.090371609 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 6 |
Hb_001247_230 |
0.1031960626 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_002849_080 |
0.1037203116 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 8 |
Hb_001301_230 |
0.105010764 |
- |
- |
PREDICTED: uncharacterized protein LOC105632949 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002609_010 |
0.1064452317 |
- |
- |
PREDICTED: uncharacterized protein LOC105630866 [Jatropha curcas] |
| 10 |
Hb_004899_070 |
0.1067734425 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 11 |
Hb_010584_010 |
0.109698259 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 12 |
Hb_003710_040 |
0.1108485349 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_003157_010 |
0.1124216751 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 14 |
Hb_000640_200 |
0.1129078404 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 15 |
Hb_000181_360 |
0.1142863031 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 16 |
Hb_057451_010 |
0.1144063242 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 17 |
Hb_007257_050 |
0.1161891858 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000813_100 |
0.1178862396 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006501_020 |
0.1193663778 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 20 |
Hb_000808_070 |
0.1194615478 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |