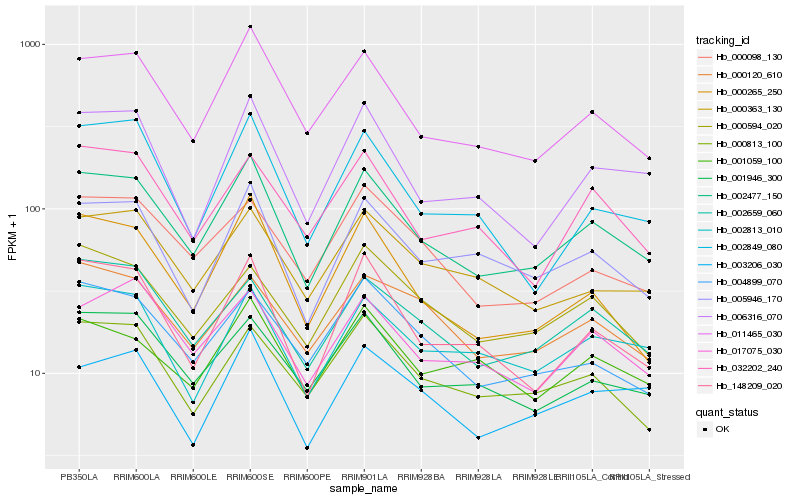
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_150 |
0.0 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_011465_030 |
0.0647924731 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_148209_020 |
0.0775039034 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 4 |
Hb_005946_170 |
0.082484556 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 5 |
Hb_000265_250 |
0.083031228 |
- |
- |
auxilin, putative [Ricinus communis] |
| 6 |
Hb_002659_060 |
0.0835164999 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 7 |
Hb_001946_300 |
0.0856017443 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 8 |
Hb_006316_070 |
0.0878884129 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 9 |
Hb_000363_130 |
0.0893919291 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 10 |
Hb_002813_010 |
0.0900923514 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 11 |
Hb_017075_030 |
0.0906144191 |
- |
- |
helicase, putative [Ricinus communis] |
| 12 |
Hb_000813_100 |
0.0910067623 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000098_130 |
0.0927248813 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 14 |
Hb_004899_070 |
0.0927854051 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 15 |
Hb_000594_020 |
0.0944375496 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 16 |
Hb_032202_240 |
0.0944416848 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 17 |
Hb_001059_100 |
0.096239129 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 18 |
Hb_003206_030 |
0.0979732912 |
- |
- |
PREDICTED: uncharacterized protein LOC105632652 [Jatropha curcas] |
| 19 |
Hb_002849_080 |
0.0988253996 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 20 |
Hb_000120_610 |
0.0993173751 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |