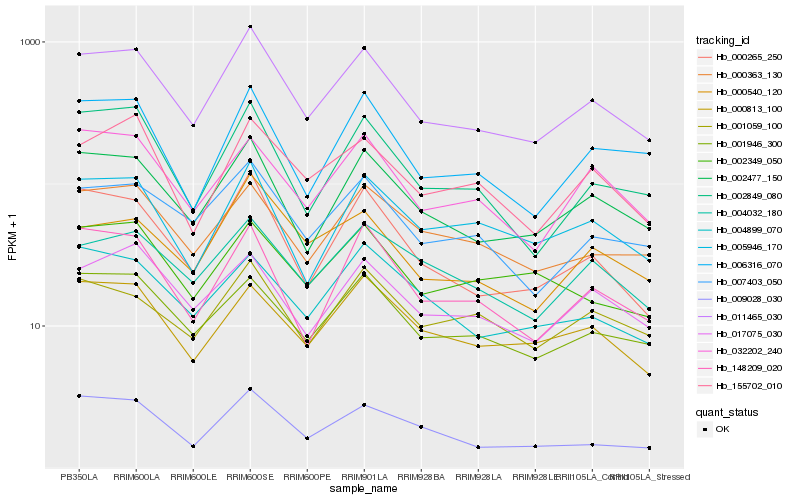
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011465_030 |
0.0 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 2 |
Hb_002477_150 |
0.0647924731 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000265_250 |
0.0743271987 |
- |
- |
auxilin, putative [Ricinus communis] |
| 4 |
Hb_007403_050 |
0.0827894181 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 5 |
Hb_001946_300 |
0.0861928452 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 6 |
Hb_002849_080 |
0.0889147897 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 7 |
Hb_000540_120 |
0.090893144 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 8 |
Hb_009028_030 |
0.0933825527 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 9 |
Hb_148209_020 |
0.0937021186 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 10 |
Hb_006316_070 |
0.0941820613 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 11 |
Hb_000363_130 |
0.0949400432 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 12 |
Hb_032202_240 |
0.09561891 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 13 |
Hb_017075_030 |
0.0963653281 |
- |
- |
helicase, putative [Ricinus communis] |
| 14 |
Hb_005946_170 |
0.0977809135 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 15 |
Hb_000813_100 |
0.0980762041 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 16 |
Hb_155702_010 |
0.0984342518 |
- |
- |
Tetraspanin-5 [Gossypium arboreum] |
| 17 |
Hb_004032_180 |
0.0998556278 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 18 |
Hb_001059_100 |
0.1054272176 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 19 |
Hb_004899_070 |
0.1054554691 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 20 |
Hb_002349_050 |
0.1074995703 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |