| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
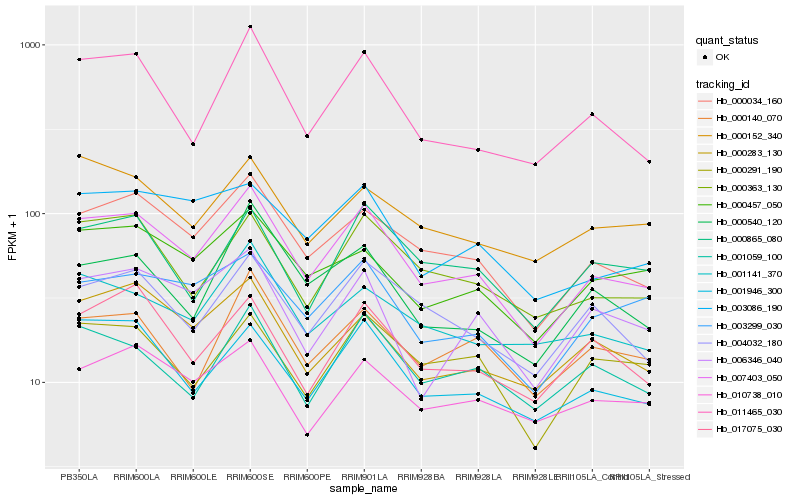
Hb_007403_050 |
0.0 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 2 |
Hb_000034_160 |
0.0619001639 |
- |
- |
hypothetical protein EUGRSUZ_F02151 [Eucalyptus grandis] |
| 3 |
Hb_011465_030 |
0.0827894181 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 4 |
Hb_001946_300 |
0.0841568853 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 5 |
Hb_000363_130 |
0.0881517887 |
- |
- |
PREDICTED: FRIGIDA-like protein 1 [Jatropha curcas] |
| 6 |
Hb_004032_180 |
0.0909448904 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 7 |
Hb_000283_130 |
0.0928200488 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 8 |
Hb_000457_050 |
0.0950326633 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 9 |
Hb_001059_100 |
0.0961519725 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 10 |
Hb_003299_030 |
0.0976706772 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000540_120 |
0.0979923442 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 12 |
Hb_003086_190 |
0.0983971324 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 13 |
Hb_017075_030 |
0.0987440095 |
- |
- |
helicase, putative [Ricinus communis] |
| 14 |
Hb_001141_370 |
0.1002004638 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 15 |
Hb_006346_040 |
0.1008978754 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 16 |
Hb_000865_080 |
0.1015499243 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 17 |
Hb_010738_010 |
0.1018917919 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 18 |
Hb_000291_190 |
0.1020709623 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 19 |
Hb_000140_070 |
0.1020720771 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000152_340 |
0.1025444449 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |