| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
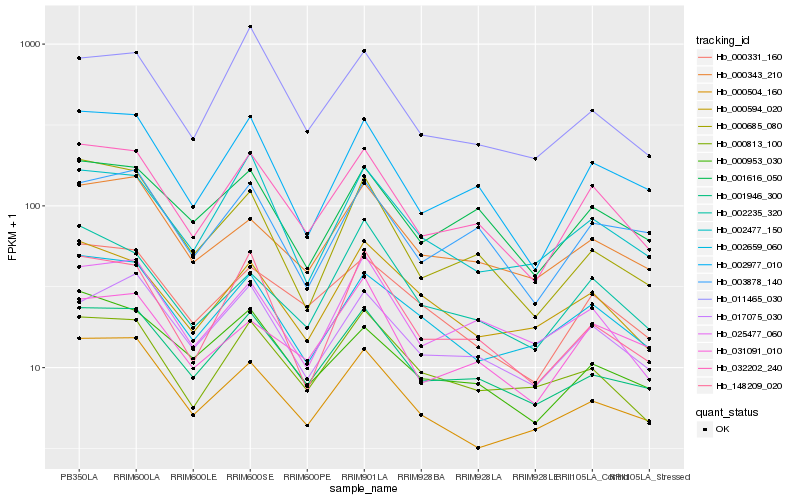
Hb_032202_240 |
0.0 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 2 |
Hb_002977_010 |
0.0641241096 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 3 |
Hb_001946_300 |
0.0720298816 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 4 |
Hb_001616_050 |
0.078654247 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 5 |
Hb_000331_160 |
0.083013046 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 6 |
Hb_017075_030 |
0.084238596 |
- |
- |
helicase, putative [Ricinus communis] |
| 7 |
Hb_025477_060 |
0.0889584393 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 8 |
Hb_000953_030 |
0.0899943553 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_003878_140 |
0.0903494743 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 10 |
Hb_000813_100 |
0.0907794049 |
- |
- |
PREDICTED: uncharacterized protein LOC105631583 isoform X2 [Jatropha curcas] |
| 11 |
Hb_148209_020 |
0.0911342906 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 12 |
Hb_002659_060 |
0.0927471378 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 13 |
Hb_002477_150 |
0.0944416848 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000343_210 |
0.0950323412 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 15 |
Hb_011465_030 |
0.09561891 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 16 |
Hb_000504_160 |
0.0965750339 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 17 |
Hb_000685_080 |
0.0976355736 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_002235_320 |
0.0981789977 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000594_020 |
0.0984393902 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 20 |
Hb_031091_010 |
0.0985148786 |
- |
- |
crooked neck protein, putative [Ricinus communis] |