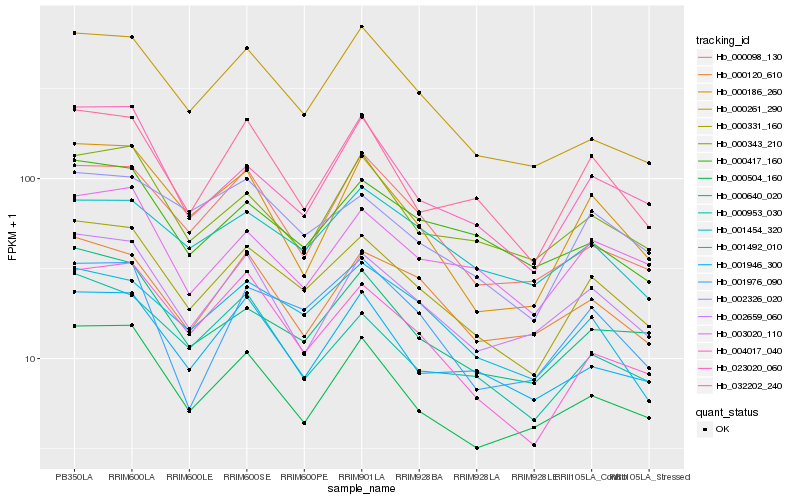
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_160 |
0.0 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 2 |
Hb_000640_020 |
0.0812018424 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 3 |
Hb_032202_240 |
0.083013046 |
- |
- |
PREDICTED: F-box protein At4g00755 [Jatropha curcas] |
| 4 |
Hb_002326_020 |
0.0837147532 |
- |
- |
hypothetical protein POPTR_0002s13160g [Populus trichocarpa] |
| 5 |
Hb_023020_060 |
0.0858503068 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 6 |
Hb_001492_010 |
0.0861780273 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like isoform X2 [Populus euphratica] |
| 7 |
Hb_000504_160 |
0.0869990186 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 8 |
Hb_004017_040 |
0.0887012672 |
- |
- |
kinetochore protein nuf2, putative [Ricinus communis] |
| 9 |
Hb_000953_030 |
0.0891674985 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_001946_300 |
0.0898764154 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 11 |
Hb_002659_060 |
0.0901343741 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 12 |
Hb_000343_210 |
0.0912601746 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 13 |
Hb_000120_610 |
0.0919752693 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 14 |
Hb_003020_110 |
0.0922978419 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 15 |
Hb_000261_290 |
0.0928080248 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 16 |
Hb_000098_130 |
0.0934040556 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 17 |
Hb_000186_260 |
0.0941696606 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 18 |
Hb_000417_160 |
0.0948684517 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 19 |
Hb_001976_090 |
0.0963403529 |
- |
- |
PREDICTED: uncharacterized protein LOC105641696 [Jatropha curcas] |
| 20 |
Hb_001454_320 |
0.0972270053 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |