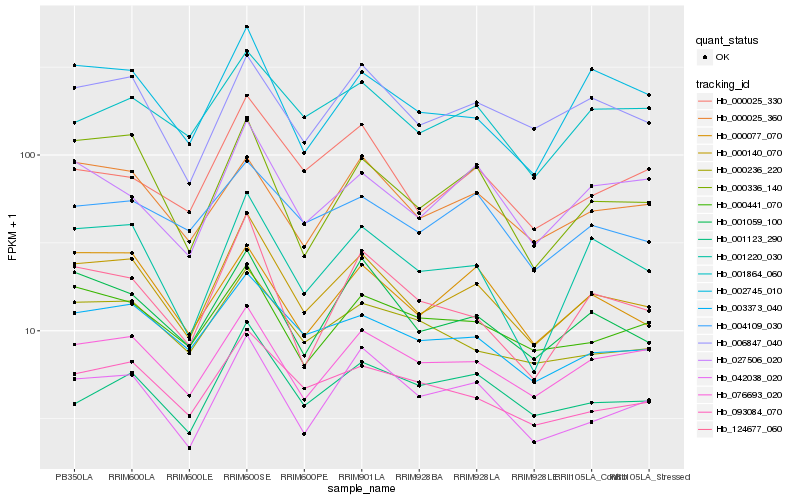
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001059_100 |
0.0804926174 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 3 |
Hb_003373_040 |
0.0828189184 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 4 |
Hb_076693_020 |
0.0870049725 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 5 |
Hb_001220_030 |
0.0874444058 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 6 |
Hb_093084_070 |
0.0880711558 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 7 |
Hb_006847_040 |
0.0900876471 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 1 [Jatropha curcas] |
| 8 |
Hb_042038_020 |
0.0923535202 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000336_140 |
0.0925512791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002745_010 |
0.0947491638 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 11 |
Hb_124677_060 |
0.0959781873 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000236_220 |
0.0969178869 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 13 |
Hb_000077_070 |
0.0970890433 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000441_070 |
0.0986529622 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000025_360 |
0.0986794194 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004109_030 |
0.09929324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001123_290 |
0.0999395298 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 3 [Jatropha curcas] |
| 18 |
Hb_001864_060 |
0.1004940339 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 19 |
Hb_027506_020 |
0.1005758853 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 20 |
Hb_000025_330 |
0.1007463887 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |