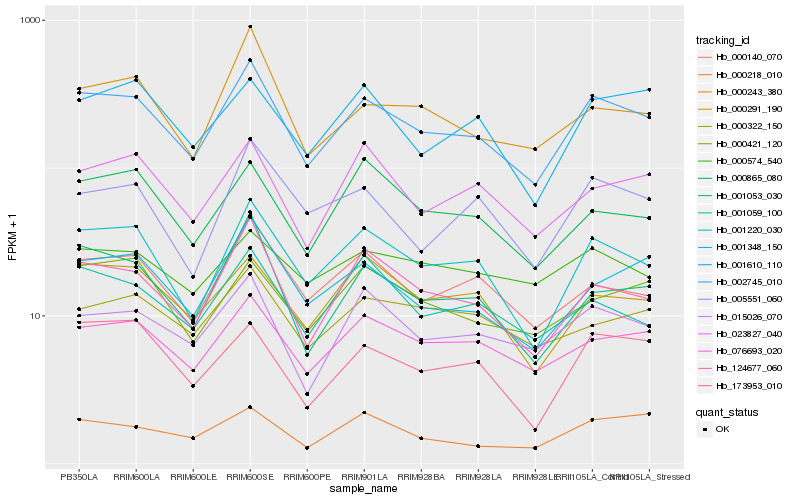
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002745_010 |
0.0 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 2 |
Hb_001220_030 |
0.0719692728 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 3 |
Hb_076693_020 |
0.0810161556 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 4 |
Hb_124677_060 |
0.0878746168 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_015026_070 |
0.0945324486 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000140_070 |
0.0947491638 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001348_150 |
0.0964343248 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001053_030 |
0.1005472477 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000291_190 |
0.1039092487 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 10 |
Hb_023827_040 |
0.106604333 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 11 |
Hb_173953_010 |
0.1077876837 |
- |
- |
hypothetical protein RCOM_1267370 [Ricinus communis] |
| 12 |
Hb_001059_100 |
0.1081662758 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000421_120 |
0.1081820249 |
- |
- |
PREDICTED: NEDD8-specific protease 1 [Jatropha curcas] |
| 14 |
Hb_000243_380 |
0.1084500307 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 15 |
Hb_000865_080 |
0.1086873893 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 16 |
Hb_005551_060 |
0.1088460024 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 17 |
Hb_000322_150 |
0.1095877074 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001610_110 |
0.1107142574 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 32 homolog 2 [Jatropha curcas] |
| 19 |
Hb_000218_010 |
0.1115464547 |
- |
- |
- |
| 20 |
Hb_000574_540 |
0.1124304295 |
- |
- |
ATP binding protein, putative [Ricinus communis] |