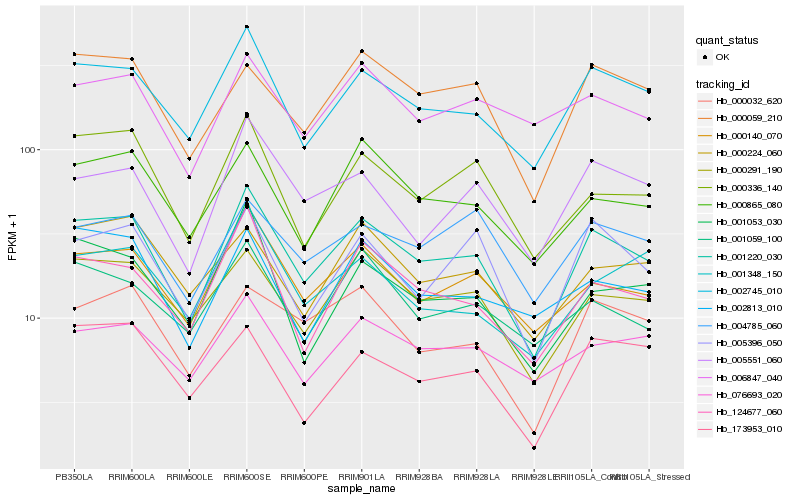
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001220_030 |
0.0 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 2 |
Hb_002745_010 |
0.0719692728 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 3 |
Hb_000140_070 |
0.0874444058 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000291_190 |
0.0923483538 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 5 |
Hb_124677_060 |
0.0970155124 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000865_080 |
0.101059518 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 7 |
Hb_000059_210 |
0.1011388292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000336_140 |
0.102793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005551_060 |
0.1031886613 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 10 |
Hb_000032_620 |
0.1038708105 |
- |
- |
hypothetical protein RCOM_1311830 [Ricinus communis] |
| 11 |
Hb_005396_050 |
0.105837252 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 12 |
Hb_076693_020 |
0.1077053893 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 13 |
Hb_001059_100 |
0.1132590377 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 14 |
Hb_001053_030 |
0.1133556556 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004785_060 |
0.1144674147 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 16 |
Hb_000224_060 |
0.1151246529 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 17 |
Hb_006847_040 |
0.1167927905 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 1 [Jatropha curcas] |
| 18 |
Hb_002813_010 |
0.1170158557 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 19 |
Hb_173953_010 |
0.1183290296 |
- |
- |
hypothetical protein RCOM_1267370 [Ricinus communis] |
| 20 |
Hb_001348_150 |
0.1189006403 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |