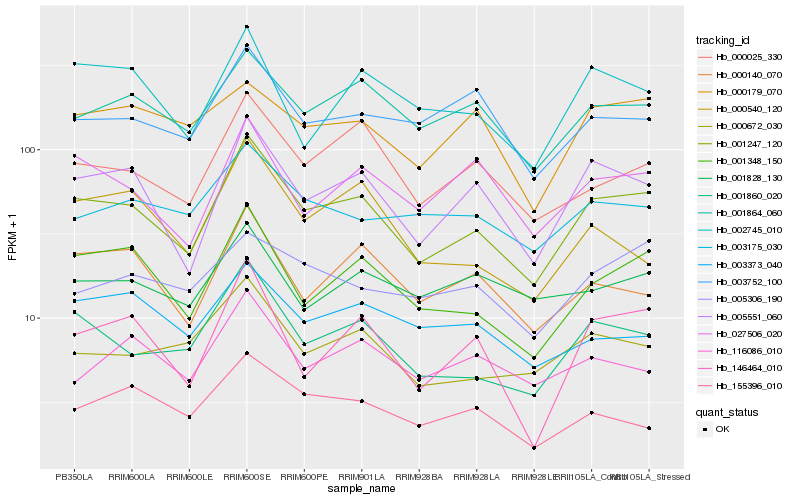
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_120 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 2 |
Hb_005551_060 |
0.0840987849 |
- |
- |
PREDICTED: uncharacterized protein LOC105635819 [Jatropha curcas] |
| 3 |
Hb_001860_020 |
0.0878322493 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 4 |
Hb_001348_150 |
0.0890651918 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001864_060 |
0.1040997321 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 6 |
Hb_000025_330 |
0.1062638803 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 7 |
Hb_000672_030 |
0.1105396004 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X1 [Jatropha curcas] |
| 8 |
Hb_027506_020 |
0.1151098023 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 9 |
Hb_146464_010 |
0.1153550874 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000140_070 |
0.1164056321 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002745_010 |
0.1169501839 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 12 |
Hb_003175_030 |
0.1220913102 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 13 |
Hb_003752_100 |
0.124865086 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 14 |
Hb_116086_010 |
0.1279062944 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 15 |
Hb_155396_010 |
0.127990965 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 16 |
Hb_001828_130 |
0.128245166 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 17 |
Hb_000179_070 |
0.1301859609 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 18 |
Hb_005306_190 |
0.130609458 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 19 |
Hb_000540_120 |
0.1310973818 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 20 |
Hb_003373_040 |
0.1320407338 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |