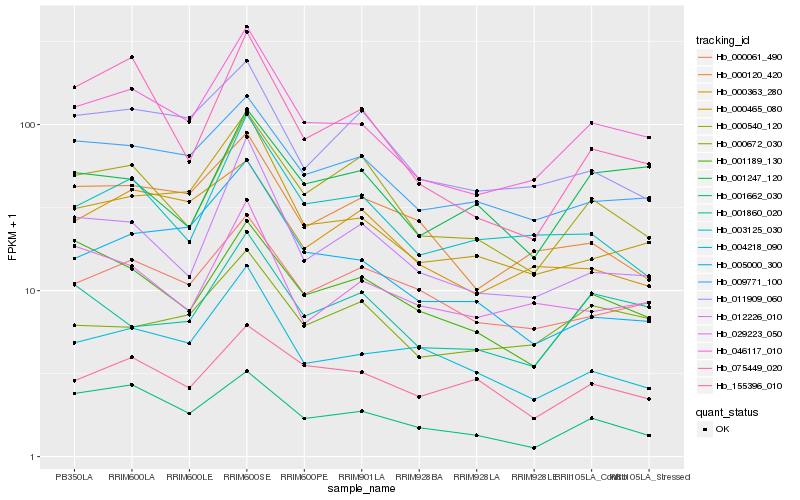
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046117_010 |
0.0 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 2 |
Hb_000465_080 |
0.0992925043 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 3 |
Hb_009771_100 |
0.1082428996 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 4 |
Hb_012226_010 |
0.1113537398 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 5 |
Hb_000540_120 |
0.1175732999 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 6 |
Hb_003125_030 |
0.1192578981 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_000061_490 |
0.1224566396 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 8 |
Hb_004218_090 |
0.1252305163 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001860_020 |
0.1259997817 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 10 |
Hb_011909_060 |
0.1268729518 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 11 |
Hb_000120_420 |
0.1292125562 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 12 |
Hb_000672_030 |
0.130277653 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001189_130 |
0.1330587754 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 14 |
Hb_001662_030 |
0.1336049388 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |
| 15 |
Hb_029223_050 |
0.133931897 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 16 |
Hb_075449_020 |
0.1366552345 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 17 |
Hb_000363_280 |
0.1367582204 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 18 |
Hb_001247_120 |
0.1395452267 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 19 |
Hb_005000_300 |
0.1397758952 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 20 |
Hb_155396_010 |
0.1409404276 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |