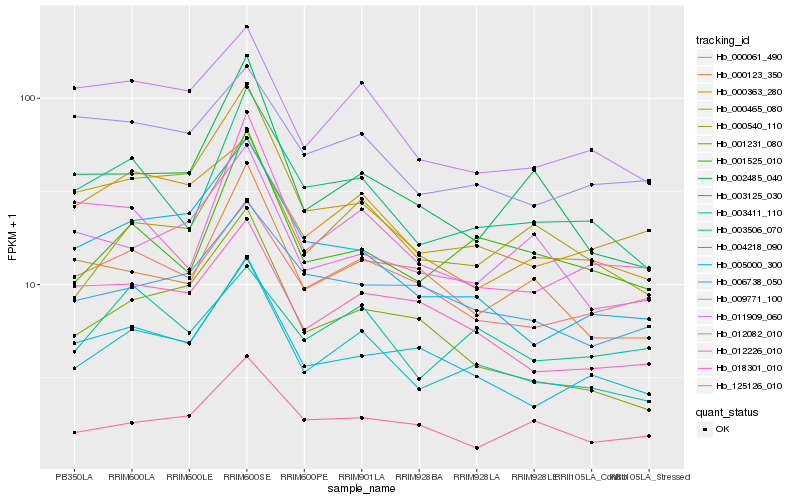
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003506_070 |
0.0 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000465_080 |
0.1007365317 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 3 |
Hb_003125_030 |
0.1015966714 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 4 |
Hb_000540_110 |
0.1062729892 |
- |
- |
hypothetical protein PRUPE_ppa000777mg [Prunus persica] |
| 5 |
Hb_001525_010 |
0.1110759895 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 6 |
Hb_011909_060 |
0.112649554 |
- |
- |
PREDICTED: zinc finger protein CONSTANS-LIKE 1 [Jatropha curcas] |
| 7 |
Hb_004218_090 |
0.1145174496 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_005000_300 |
0.1199544108 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 9 |
Hb_018301_010 |
0.1233550923 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 10 |
Hb_000061_490 |
0.1245561551 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 11 |
Hb_000363_280 |
0.1265362063 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: angiomotin [Cucumis sativus] |
| 12 |
Hb_002485_040 |
0.1267342472 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 13 |
Hb_000123_350 |
0.1281426514 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_006738_050 |
0.1285717498 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 15 |
Hb_009771_100 |
0.1295133621 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 16 |
Hb_001231_080 |
0.1304095567 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_003411_110 |
0.1307912972 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 18 |
Hb_012082_010 |
0.1308781379 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_012226_010 |
0.1337281622 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 20 |
Hb_125126_010 |
0.1350189069 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |