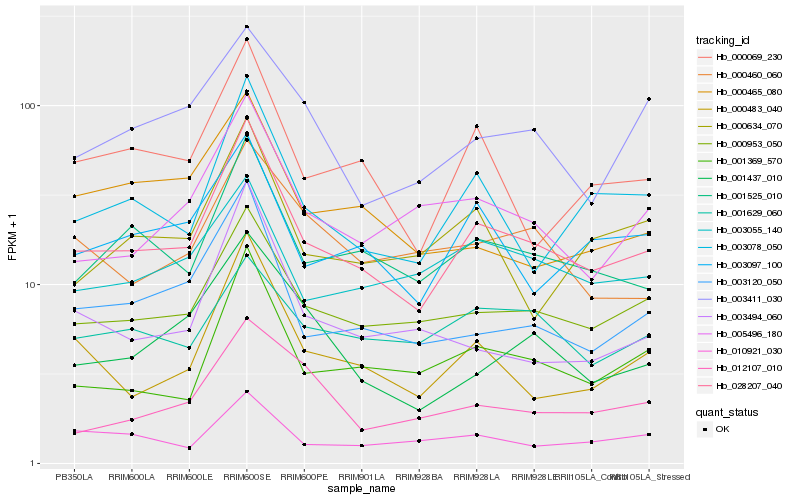
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028207_040 |
0.0 |
- |
- |
PREDICTED: beta-hexosaminidase 2 [Jatropha curcas] |
| 2 |
Hb_000953_050 |
0.1000890847 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 3 |
Hb_001525_010 |
0.1056652864 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 4 |
Hb_003078_050 |
0.1170458485 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 5 |
Hb_003120_050 |
0.1193170741 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 6 |
Hb_001369_570 |
0.1198966381 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 7 |
Hb_000069_230 |
0.1213282224 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 8 |
Hb_000483_040 |
0.1235621276 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 9 |
Hb_003097_100 |
0.1259461625 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 10 |
Hb_005496_180 |
0.1339120615 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 11 |
Hb_003055_140 |
0.1391494931 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 12 |
Hb_000634_070 |
0.1428273355 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 13 |
Hb_010921_030 |
0.1434740418 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At2g01810 [Jatropha curcas] |
| 14 |
Hb_001437_010 |
0.1459090532 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 15 |
Hb_000460_060 |
0.1465549477 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 16 |
Hb_003411_030 |
0.1470734735 |
- |
- |
hypothetical protein POPTR_0010s13590g [Populus trichocarpa] |
| 17 |
Hb_003494_060 |
0.1516609168 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001629_060 |
0.1519225627 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
Flavin-containing amine oxidase domain-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000465_080 |
0.1558852783 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 20 |
Hb_012107_010 |
0.1565171919 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |