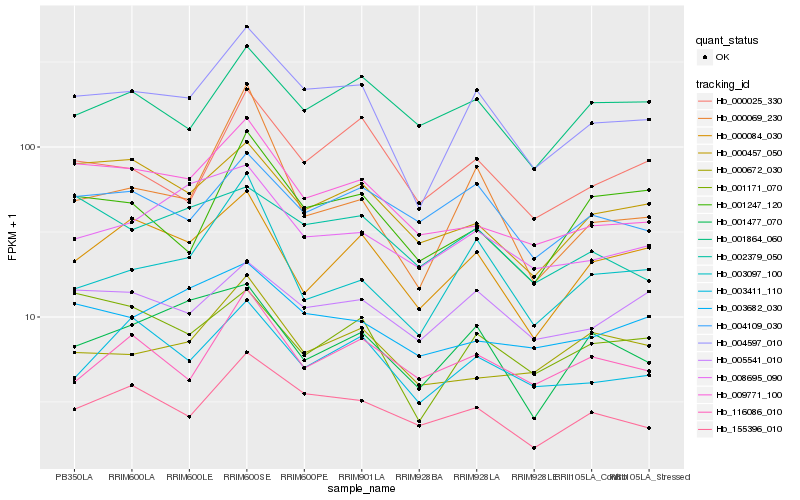
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004597_010 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MYC2 [Jatropha curcas] |
| 2 |
Hb_155396_010 |
0.1059765445 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 3 |
Hb_009771_100 |
0.1185610042 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 4 |
Hb_003411_110 |
0.1211815459 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 5 |
Hb_000025_330 |
0.1219379439 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 6 |
Hb_000084_030 |
0.1264669485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005541_010 |
0.1270326614 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 8 |
Hb_004109_030 |
0.1285521927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003682_030 |
0.1307793838 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 10 |
Hb_008695_090 |
0.1320312434 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 11 |
Hb_001171_070 |
0.1321739612 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003097_100 |
0.1325910278 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 13 |
Hb_001864_060 |
0.1326210678 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 14 |
Hb_001477_070 |
0.1329987165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001247_120 |
0.1343168415 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 3 [Jatropha curcas] |
| 16 |
Hb_000457_050 |
0.1354913582 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 17 |
Hb_116086_010 |
0.1365681449 |
- |
- |
hypothetical protein JCGZ_14829 [Jatropha curcas] |
| 18 |
Hb_000672_030 |
0.1370257564 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002379_050 |
0.1372721123 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 20 |
Hb_000069_230 |
0.1373038772 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |