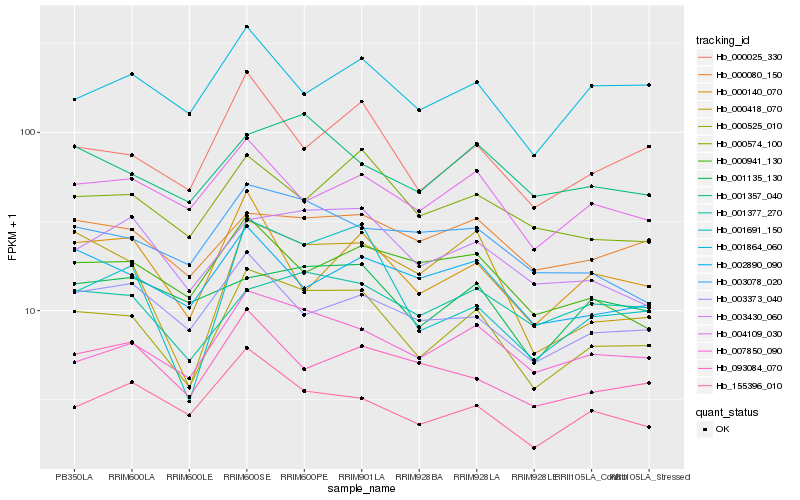
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000525_010 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_007850_090 |
0.1008437099 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000025_330 |
0.1082967071 |
- |
- |
hypothetical protein JCGZ_22759 [Jatropha curcas] |
| 4 |
Hb_001691_150 |
0.1119805235 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Jatropha curcas] |
| 5 |
Hb_000418_070 |
0.1147781541 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 6 |
Hb_003430_060 |
0.1164647545 |
- |
- |
PREDICTED: potassium transporter 23 isoform X2 [Elaeis guineensis] |
| 7 |
Hb_000574_100 |
0.1200602731 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 8 |
Hb_155396_010 |
0.1217072071 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 9 |
Hb_000080_150 |
0.122347135 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004109_030 |
0.1230716162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001357_040 |
0.1243826774 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 12 |
Hb_003078_020 |
0.1261354939 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 13 |
Hb_001864_060 |
0.1261652465 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 14 |
Hb_003373_040 |
0.1271591768 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_002890_090 |
0.1272947393 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 16 |
Hb_001135_130 |
0.1275335186 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 17 |
Hb_093084_070 |
0.1292947676 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 18 |
Hb_000140_070 |
0.1298982028 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000941_130 |
0.1300807271 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 20 |
Hb_001377_270 |
0.1304228048 |
- |
- |
hypothetical protein CICLE_v10021159mg [Citrus clementina] |