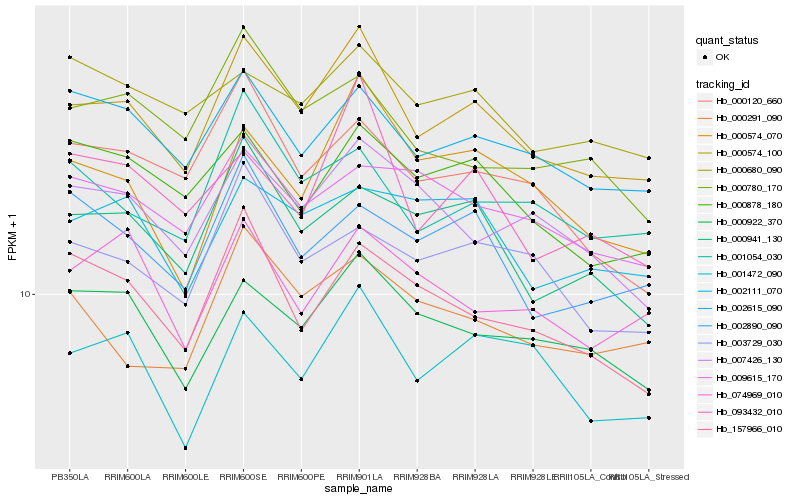
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_100 |
0.0 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 2 |
Hb_002615_090 |
0.0779778609 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000922_370 |
0.0794029883 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 4 |
Hb_001472_090 |
0.0839251901 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000941_130 |
0.0845678493 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 6 |
Hb_000574_070 |
0.0856835686 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 7 |
Hb_000878_180 |
0.0867531988 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007426_130 |
0.0877973801 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 9 |
Hb_093432_010 |
0.0899861419 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 10 |
Hb_001054_030 |
0.0902280888 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 11 |
Hb_003729_030 |
0.0924733546 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 12 |
Hb_000120_660 |
0.0928049631 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 13 |
Hb_157966_010 |
0.0930068881 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 14 |
Hb_002890_090 |
0.0948009999 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 15 |
Hb_009615_170 |
0.0949272387 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002111_070 |
0.0951831959 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 17 |
Hb_000291_090 |
0.0951949776 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000780_170 |
0.0952667823 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 19 |
Hb_000680_090 |
0.0953812289 |
- |
- |
Protein SEY1, putative [Ricinus communis] |
| 20 |
Hb_074969_010 |
0.0954782208 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |