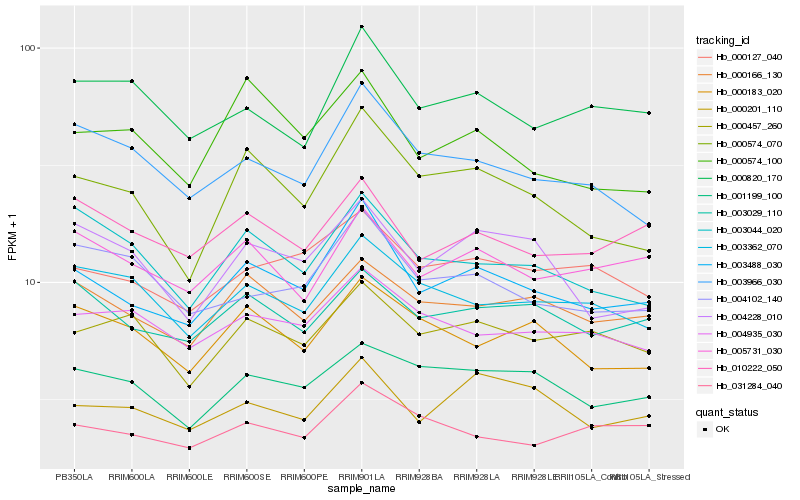
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003488_030 |
0.0 |
- |
- |
PREDICTED: protein CPR-5 [Jatropha curcas] |
| 2 |
Hb_000574_070 |
0.0809659245 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 3 |
Hb_000127_040 |
0.0815471732 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 4 |
Hb_000166_130 |
0.0841281741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003029_110 |
0.0860763914 |
- |
- |
PREDICTED: uncharacterized protein LOC105640062 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000201_110 |
0.0882806657 |
- |
- |
hypothetical protein JCGZ_21690 [Jatropha curcas] |
| 7 |
Hb_003966_030 |
0.0885837449 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 8 |
Hb_005731_030 |
0.0886499855 |
- |
- |
PREDICTED: UPF0400 protein C337.03 [Jatropha curcas] |
| 9 |
Hb_004935_030 |
0.0909768669 |
- |
- |
PREDICTED: uncharacterized protein LOC102628125 [Citrus sinensis] |
| 10 |
Hb_010222_050 |
0.0918683601 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000820_170 |
0.092705706 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 12 |
Hb_000457_260 |
0.0930197449 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15690-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_031284_040 |
0.093126662 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g30700 [Jatropha curcas] |
| 14 |
Hb_001199_100 |
0.0940655412 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 15 |
Hb_003362_070 |
0.0943288898 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 16 |
Hb_000183_020 |
0.0950972253 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004102_140 |
0.0962845612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003044_020 |
0.0964315164 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 19 |
Hb_000574_100 |
0.0966733467 |
- |
- |
DEAD-box protein abstrakt [Populus trichocarpa] |
| 20 |
Hb_004228_010 |
0.0966932914 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA [Jatropha curcas] |