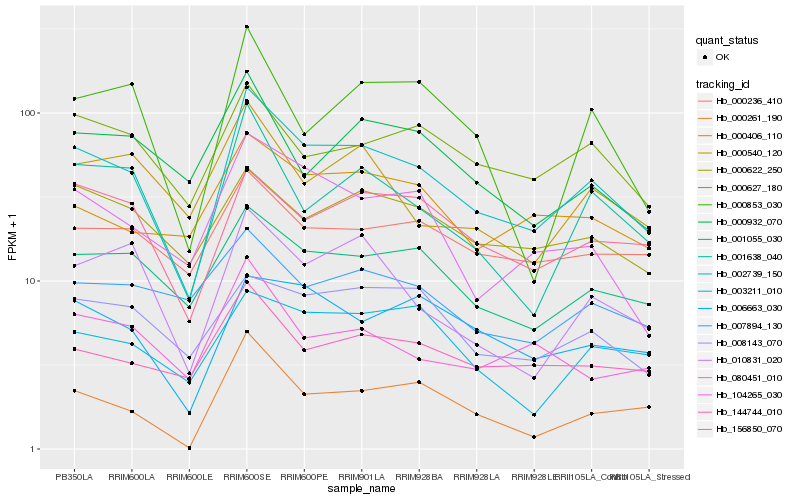
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002739_150 |
0.0 |
- |
- |
Uncharacterized protein TCM_016791 [Theobroma cacao] |
| 2 |
Hb_001055_030 |
0.1210977115 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001638_040 |
0.123171571 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 4 |
Hb_156850_070 |
0.1286197716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_006663_030 |
0.1339729483 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 6 |
Hb_080451_010 |
0.1343217115 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 7 |
Hb_010831_020 |
0.1351189645 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 8 |
Hb_144744_010 |
0.135832832 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 9 |
Hb_000627_180 |
0.1372645086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000622_250 |
0.1400553969 |
- |
- |
atpob1, putative [Ricinus communis] |
| 11 |
Hb_008143_070 |
0.1411305327 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 12 |
Hb_000236_410 |
0.1422037391 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 13 |
Hb_000261_190 |
0.1433490056 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 14 |
Hb_000406_110 |
0.1435884915 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 [Jatropha curcas] |
| 15 |
Hb_000540_120 |
0.1471715391 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 16 |
Hb_104265_030 |
0.1492540299 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 17 |
Hb_003211_010 |
0.1495241598 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 18 |
Hb_000932_070 |
0.1496756049 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 19 |
Hb_000853_030 |
0.1507164014 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 20 |
Hb_007894_130 |
0.1511179313 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |