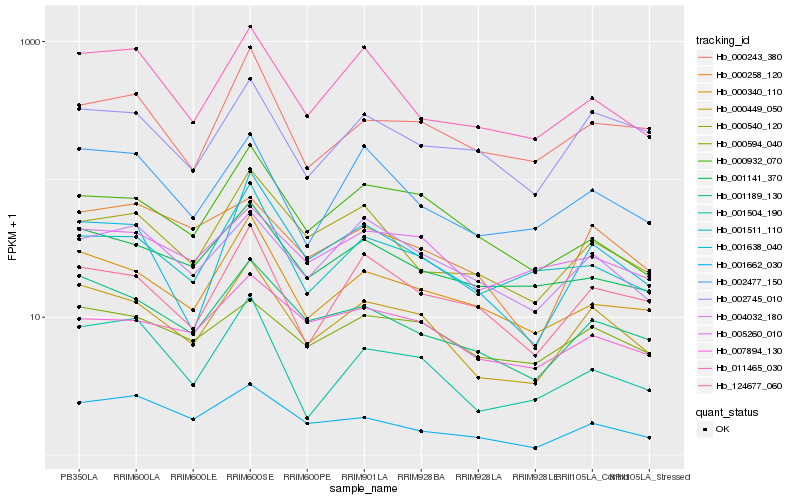
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000449_050 |
0.0 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 2 |
Hb_001189_130 |
0.091072735 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 3 |
Hb_000340_110 |
0.1097200631 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 4 |
Hb_001638_040 |
0.1129509709 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 5 |
Hb_002477_150 |
0.1185671559 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_124677_060 |
0.1202788073 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_004032_180 |
0.1220220696 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 8 |
Hb_001511_110 |
0.1222557533 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001141_370 |
0.1237494125 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 10 |
Hb_011465_030 |
0.1239228592 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_002745_010 |
0.124998481 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 12 |
Hb_000540_120 |
0.1254296008 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 13 |
Hb_000932_070 |
0.1270716325 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 14 |
Hb_000594_040 |
0.1272903497 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 15 |
Hb_007894_130 |
0.129512341 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 16 |
Hb_001504_190 |
0.1307692209 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_005260_010 |
0.1310209219 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000243_380 |
0.131852233 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 19 |
Hb_000258_120 |
0.1327892716 |
- |
- |
PREDICTED: uncharacterized protein LOC105630440 [Jatropha curcas] |
| 20 |
Hb_001662_030 |
0.1340496338 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Populus euphratica] |