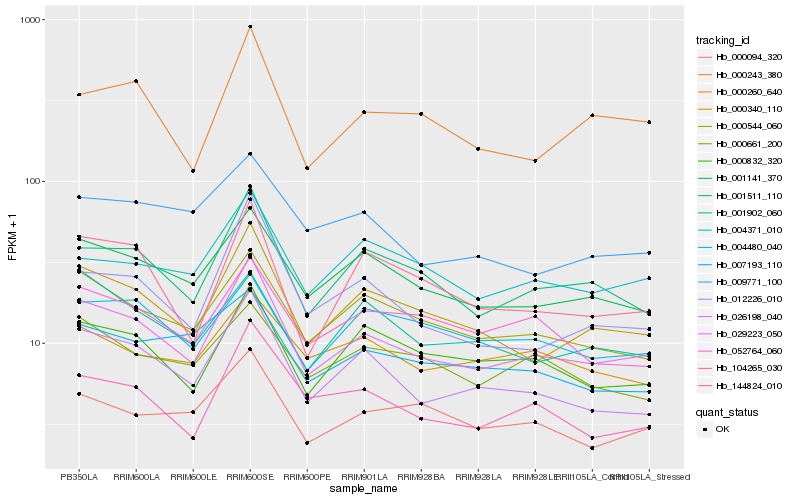
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029223_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000340_110 |
0.069707 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 3 |
Hb_001511_110 |
0.0777766166 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_026198_040 |
0.0797483918 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 5 |
Hb_001141_370 |
0.0868392712 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 6 |
Hb_000544_060 |
0.0916020848 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 7 |
Hb_104265_030 |
0.0943061467 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 8 |
Hb_000832_320 |
0.0949315629 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000243_380 |
0.0952326742 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 10 |
Hb_012226_010 |
0.1009690054 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 11 |
Hb_144824_010 |
0.1016068896 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 12 |
Hb_000094_320 |
0.1046768509 |
- |
- |
hypothetical protein JCGZ_04407 [Jatropha curcas] |
| 13 |
Hb_004371_010 |
0.10680573 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_052764_060 |
0.1087704952 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 15 |
Hb_004480_040 |
0.115428361 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000260_640 |
0.116399119 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 17 |
Hb_009771_100 |
0.1165817072 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 18 |
Hb_007193_110 |
0.117185635 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 19 |
Hb_000661_200 |
0.1184673491 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001902_060 |
0.1189847043 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |