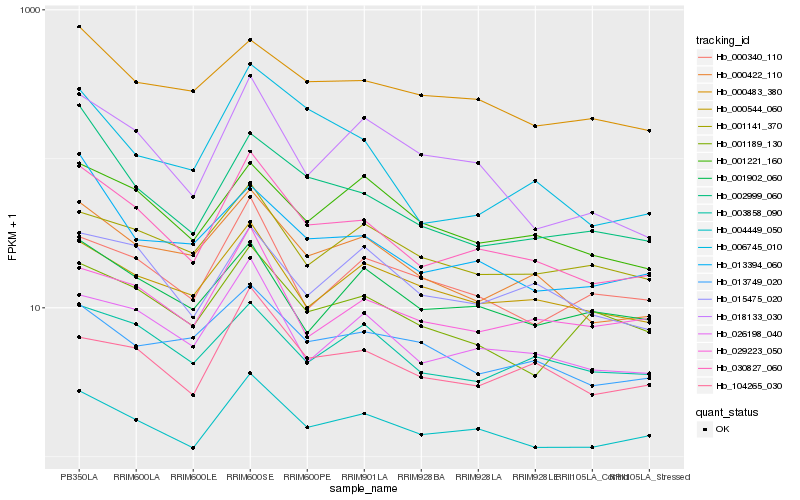
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030827_060 |
0.0 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2b [Jatropha curcas] |
| 2 |
Hb_000422_110 |
0.0949014578 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 3 |
Hb_026198_040 |
0.0991729488 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 4 |
Hb_104265_030 |
0.116585544 |
- |
- |
hypothetical protein POPTR_0001s31230g [Populus trichocarpa] |
| 5 |
Hb_018133_030 |
0.1183019444 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 6 |
Hb_004449_050 |
0.1201419844 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Jatropha curcas] |
| 7 |
Hb_002999_060 |
0.1215076549 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 8 |
Hb_000544_060 |
0.1221417674 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 9 |
Hb_029223_050 |
0.1235173508 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003858_090 |
0.1246885619 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 11 |
Hb_001221_160 |
0.1253442948 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 12 |
Hb_000483_380 |
0.1255056723 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 13 |
Hb_001189_130 |
0.1267007591 |
transcription factor |
TF Family: NAC |
PREDICTED: protein ATAF2-like [Jatropha curcas] |
| 14 |
Hb_015475_020 |
0.1275684836 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 15 |
Hb_013394_060 |
0.1277211521 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 16 |
Hb_013749_020 |
0.1287482021 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_006745_010 |
0.1288998347 |
- |
- |
PREDICTED: uncharacterized protein LOC105640095 [Jatropha curcas] |
| 18 |
Hb_000340_110 |
0.1293324717 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 19 |
Hb_001141_370 |
0.1308418143 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 20 |
Hb_001902_060 |
0.134692693 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |