| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
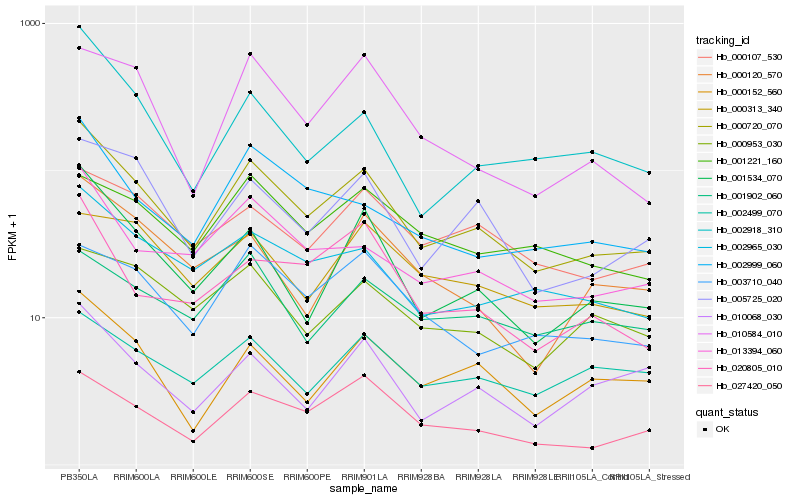
Hb_000720_070 |
0.0 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 2 |
Hb_002965_030 |
0.1091780344 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 3 |
Hb_001534_070 |
0.1109264728 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 4 |
Hb_013394_060 |
0.1141381829 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_005725_020 |
0.1171880703 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 [Jatropha curcas] |
| 6 |
Hb_027420_050 |
0.117539592 |
- |
- |
- |
| 7 |
Hb_002999_060 |
0.1230750073 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 8 |
Hb_002918_310 |
0.1252887202 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 9 |
Hb_000152_560 |
0.1272110177 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 10 |
Hb_003710_040 |
0.1305725726 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000107_530 |
0.131237676 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 12 |
Hb_020805_010 |
0.1365837533 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000953_030 |
0.1367836194 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_010584_010 |
0.1374747108 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 15 |
Hb_010068_030 |
0.1382466458 |
- |
- |
aconitase, putative [Ricinus communis] |
| 16 |
Hb_001221_160 |
0.1395264692 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 17 |
Hb_002499_070 |
0.1397702999 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000120_570 |
0.1401828357 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 19 |
Hb_001902_060 |
0.1408176746 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000313_340 |
0.1408840661 |
- |
- |
DNA repair protein xp-E, putative [Ricinus communis] |