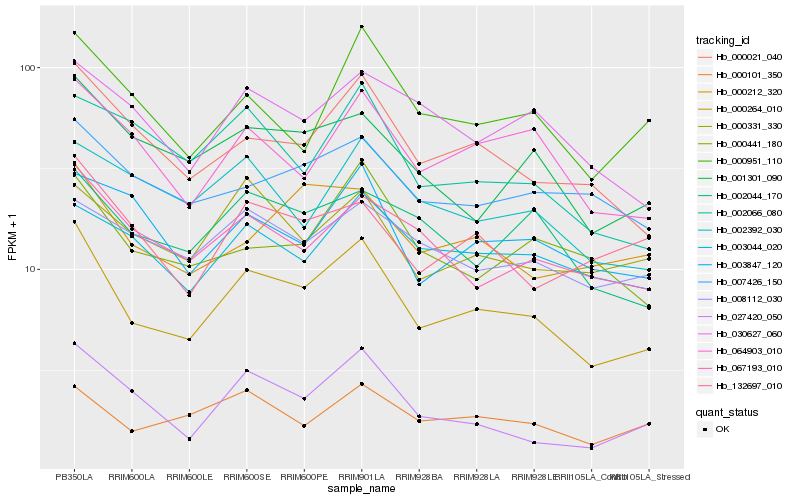
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649505 [Jatropha curcas] |
| 2 |
Hb_000021_040 |
0.1025469647 |
- |
- |
PREDICTED: villin-3-like isoform X1 [Populus euphratica] |
| 3 |
Hb_064903_010 |
0.1141600513 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 4 |
Hb_067193_010 |
0.1214255696 |
- |
- |
- |
| 5 |
Hb_001301_090 |
0.1222172926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000101_350 |
0.1237813306 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance protein, putative [Theobroma cacao] |
| 7 |
Hb_000951_110 |
0.1246816721 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 8 |
Hb_000331_330 |
0.1259480798 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002044_170 |
0.1267610099 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 10 |
Hb_000441_180 |
0.1271269477 |
- |
- |
Early endosome antigen, putative [Ricinus communis] |
| 11 |
Hb_027420_050 |
0.1272885954 |
- |
- |
- |
| 12 |
Hb_000212_320 |
0.1296837191 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 13 |
Hb_003847_120 |
0.1297255055 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 14 |
Hb_030627_060 |
0.1298214862 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 15 |
Hb_132697_010 |
0.1301176747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003044_020 |
0.1303475302 |
- |
- |
PREDICTED: uncharacterized protein LOC105641479 [Jatropha curcas] |
| 17 |
Hb_007426_150 |
0.131401639 |
- |
- |
PREDICTED: F-box protein At5g46170-like [Jatropha curcas] |
| 18 |
Hb_002392_030 |
0.1319862037 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 19 |
Hb_008112_030 |
0.1327201072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002066_080 |
0.132757854 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |