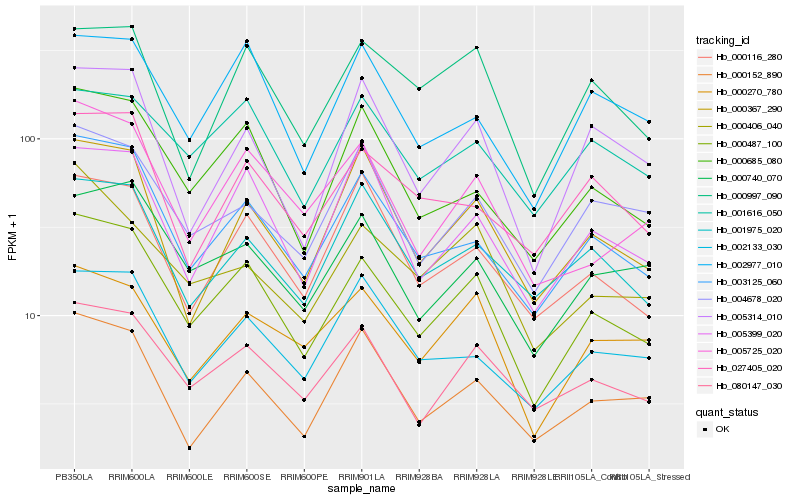
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_100 |
0.0 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 2 |
Hb_003125_060 |
0.0835222966 |
- |
- |
PREDICTED: snurportin-1 [Jatropha curcas] |
| 3 |
Hb_080147_030 |
0.0896537287 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000685_080 |
0.0950050516 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_005725_020 |
0.0998090606 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 [Jatropha curcas] |
| 6 |
Hb_000740_070 |
0.1068787729 |
- |
- |
- |
| 7 |
Hb_005399_020 |
0.1125708543 |
- |
- |
hypothetical protein CISIN_1g022820mg [Citrus sinensis] |
| 8 |
Hb_004678_020 |
0.1158829585 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 9 |
Hb_000270_780 |
0.1163930201 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 10 |
Hb_000367_290 |
0.117152022 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |
| 11 |
Hb_000152_890 |
0.1172010351 |
- |
- |
hypothetical protein JCGZ_26265 [Jatropha curcas] |
| 12 |
Hb_002977_010 |
0.1172074986 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 13 |
Hb_000116_280 |
0.1174214423 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 14 |
Hb_002133_030 |
0.1175529319 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 15 |
Hb_005314_010 |
0.1177077515 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 16 |
Hb_001975_020 |
0.1194504016 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 17 |
Hb_001616_050 |
0.1207472873 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 18 |
Hb_000406_040 |
0.1208978875 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 19 |
Hb_000997_090 |
0.1216363558 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 20 |
Hb_027405_020 |
0.1234376213 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |