| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
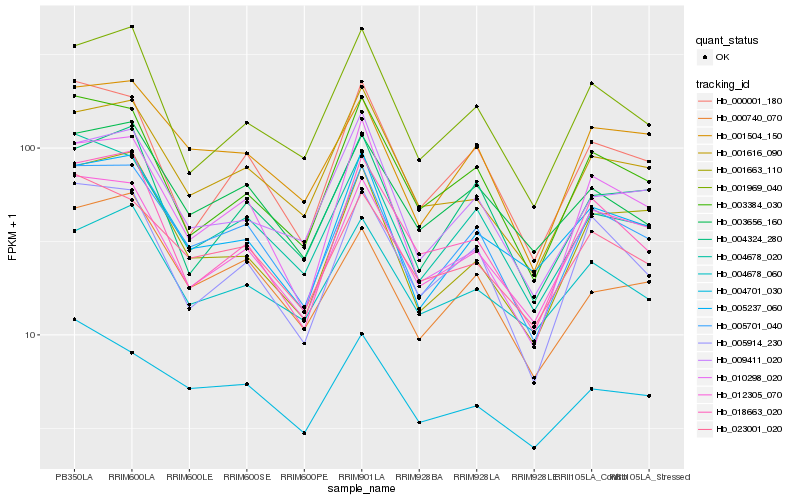
Hb_005701_040 |
0.0 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_001504_150 |
0.0608727763 |
- |
- |
PREDICTED: DCN1-like protein 4 [Jatropha curcas] |
| 3 |
Hb_004678_020 |
0.063178351 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 4 |
Hb_012305_070 |
0.0707294612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005914_230 |
0.0720583673 |
- |
- |
PREDICTED: pre-mRNA-processing factor 19 homolog 1 [Jatropha curcas] |
| 6 |
Hb_023001_020 |
0.0732881563 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 7 |
Hb_003656_160 |
0.0772050504 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein B456_013G226400 [Gossypium raimondii] |
| 8 |
Hb_005237_060 |
0.0777536853 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 9 |
Hb_000001_180 |
0.0807389232 |
- |
- |
PREDICTED: uncharacterized protein LOC105648978 [Jatropha curcas] |
| 10 |
Hb_001616_090 |
0.081321777 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003384_030 |
0.0817653777 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit beta-like [Jatropha curcas] |
| 12 |
Hb_001969_040 |
0.0851515382 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 13 |
Hb_000740_070 |
0.0865671465 |
- |
- |
- |
| 14 |
Hb_001663_110 |
0.0879486389 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_009411_020 |
0.0889875614 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 16 |
Hb_004678_060 |
0.0892610848 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 17 |
Hb_004324_280 |
0.0899433768 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 18 |
Hb_004701_030 |
0.0904071219 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 19 |
Hb_018663_020 |
0.0913785789 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_010298_020 |
0.0913919915 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 isoform X4 [Vitis vinifera] |