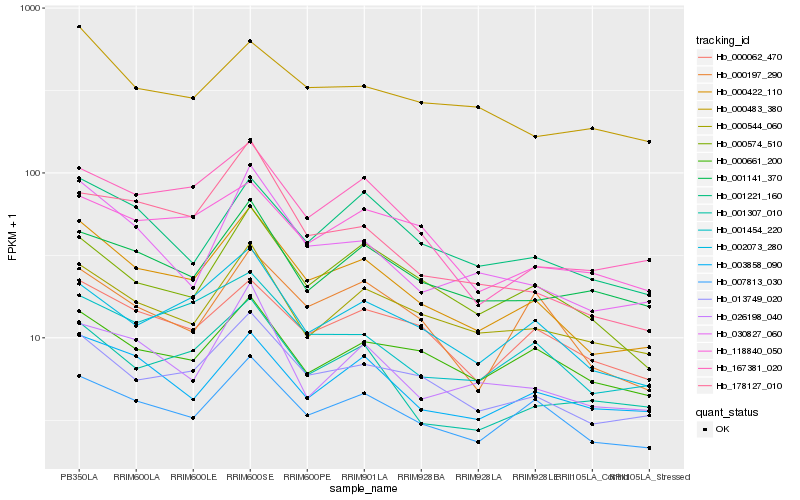
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 2 |
Hb_013749_020 |
0.0692558429 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_007813_030 |
0.0875988904 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 4 |
Hb_002073_280 |
0.0940707838 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_030827_060 |
0.0949014578 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2b [Jatropha curcas] |
| 6 |
Hb_000574_510 |
0.0972839405 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000544_060 |
0.0983092751 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 8 |
Hb_167381_020 |
0.0991234111 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 9 |
Hb_001221_160 |
0.1010103929 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 10 |
Hb_001454_220 |
0.1013447073 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 11 |
Hb_000062_470 |
0.1021877151 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 12 |
Hb_001307_010 |
0.1029404878 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 13 |
Hb_003858_090 |
0.1054709962 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 14 |
Hb_000197_290 |
0.1085403222 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 15 |
Hb_001141_370 |
0.1099554015 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 16 |
Hb_026198_040 |
0.1100086101 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 17 |
Hb_000661_200 |
0.111092473 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 18 |
Hb_118840_050 |
0.113362307 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 19 |
Hb_178127_010 |
0.1137443828 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 20 |
Hb_000483_380 |
0.1140938277 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |